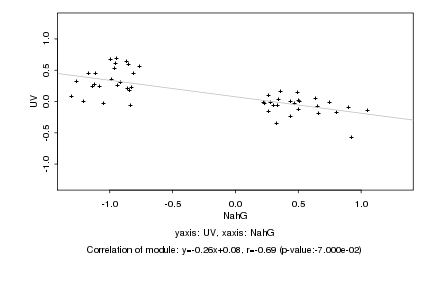
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
NahG |
Log2 signal ratio
UV |
| 1 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.047 |
-0.131 |
| 2 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.923 |
-0.570 |
| 3 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.892 |
-0.084 |
| 4 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.798 |
-0.164 |
| 5 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.741 |
-0.003 |
| 6 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.657 |
-0.185 |
| 7 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.652 |
-0.071 |
| 8 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.633 |
0.056 |
| 9 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.503 |
0.007 |
| 10 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.501 |
0.021 |
| 11 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.497 |
-0.114 |
| 12 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.493 |
0.149 |
| 13 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.462 |
-0.022 |
| 14 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.436 |
0.014 |
| 15 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.434 |
-0.235 |
| 16 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.358 |
0.163 |
| 17 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.339 |
0.040 |
| 18 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
0.328 |
-0.063 |
| 19 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.324 |
-0.347 |
| 20 |
246030_at |
AT5G21105
|
[Plant L-ascorbate oxidase] |
0.299 |
-0.048 |
| 21 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.271 |
-0.001 |
| 22 |
250020_at |
AT5G18180
|
[H/ACA ribonucleoprotein complex, subunit Gar1/Naf1 protein] |
0.260 |
0.111 |
| 23 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
0.259 |
-0.155 |
| 24 |
252995_at |
AT4G38370
|
[Phosphoglycerate mutase family protein] |
0.228 |
-0.028 |
| 25 |
263235_at |
AT1G10430
|
PP2A-2, protein phosphatase 2A-2 |
0.216 |
-0.002 |
| 26 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-1.310 |
0.088 |
| 27 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-1.268 |
0.321 |
| 28 |
261221_at |
AT1G19960
|
unknown |
-1.213 |
0.007 |
| 29 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-1.171 |
0.450 |
| 30 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-1.138 |
0.241 |
| 31 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-1.122 |
0.280 |
| 32 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-1.121 |
0.462 |
| 33 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-1.086 |
0.252 |
| 34 |
253730_at |
AT4G29480
|
[Mitochondrial ATP synthase subunit G protein] |
-1.051 |
-0.031 |
| 35 |
259385_at |
AT1G13470
|
unknown |
-1.000 |
0.672 |
| 36 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.992 |
0.365 |
| 37 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.963 |
0.529 |
| 38 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.956 |
0.617 |
| 39 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.949 |
0.694 |
| 40 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.941 |
0.258 |
| 41 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.922 |
0.313 |
| 42 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.869 |
0.644 |
| 43 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.860 |
0.210 |
| 44 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.852 |
0.604 |
| 45 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.847 |
0.178 |
| 46 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.836 |
-0.056 |
| 47 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.831 |
0.238 |
| 48 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.814 |
0.457 |
| 49 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.765 |
0.565 |