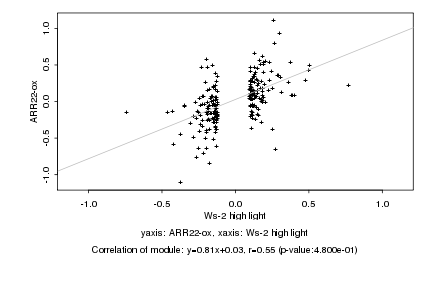
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ws-2 high light |
Log2 signal ratio
ARR22-ox |
| 1 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.765 |
0.228 |
| 2 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.504 |
0.497 |
| 3 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.491 |
0.437 |
| 4 |
267181_at |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
0.475 |
0.289 |
| 5 |
266912_at |
AT2G45900
|
TRM13, TON1 Recruiting Motif 13 |
0.396 |
0.085 |
| 6 |
258370_at |
AT3G14395
|
unknown |
0.386 |
0.085 |
| 7 |
248867_at |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
0.380 |
0.084 |
| 8 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.369 |
0.540 |
| 9 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.357 |
0.267 |
| 10 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.309 |
0.131 |
| 11 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.305 |
0.341 |
| 12 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.298 |
0.942 |
| 13 |
253940_at |
AT4G26950
|
unknown |
0.293 |
0.369 |
| 14 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.282 |
0.357 |
| 15 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.270 |
-0.643 |
| 16 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.260 |
0.801 |
| 17 |
256603_at |
AT3G28270
|
unknown |
0.257 |
1.121 |
| 18 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.252 |
-0.369 |
| 19 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.250 |
0.179 |
| 20 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.243 |
0.413 |
| 21 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.231 |
0.541 |
| 22 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.228 |
0.300 |
| 23 |
257456_at |
AT2G18120
|
SRS4, SHI-related sequence 4 |
0.220 |
0.155 |
| 24 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
0.203 |
-0.006 |
| 25 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.203 |
0.556 |
| 26 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.197 |
0.236 |
| 27 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
0.190 |
0.516 |
| 28 |
261212_at |
AT1G12880
|
atnudt12, nudix hydrolase homolog 12, NUDT12, nudix hydrolase homolog 12 |
0.189 |
0.535 |
| 29 |
246403_at |
AT1G57590
|
[Pectinacetylesterase family protein] |
0.187 |
0.397 |
| 30 |
246552_at |
AT5G15420
|
unknown |
0.187 |
0.055 |
| 31 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.184 |
0.627 |
| 32 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.183 |
0.093 |
| 33 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.181 |
0.149 |
| 34 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.180 |
-0.001 |
| 35 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
0.179 |
0.184 |
| 36 |
260146_at |
AT1G52770
|
[Phototropic-responsive NPH3 family protein] |
0.178 |
0.023 |
| 37 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.177 |
0.028 |
| 38 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.175 |
-0.281 |
| 39 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.173 |
0.277 |
| 40 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.172 |
0.011 |
| 41 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.169 |
0.187 |
| 42 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.164 |
0.517 |
| 43 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.162 |
0.565 |
| 44 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.160 |
0.047 |
| 45 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.155 |
-0.099 |
| 46 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.155 |
-0.178 |
| 47 |
246429_at |
AT5G17450
|
HIPP21, heavy metal associated isoprenylated plant protein 21 |
0.151 |
0.252 |
| 48 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
0.149 |
-0.166 |
| 49 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.147 |
0.230 |
| 50 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.146 |
0.119 |
| 51 |
257719_at |
AT3G18440
|
ALMT9, aluminum-activated malate transporter 9, AtALMT9, aluminum-activated malate transporter 9 |
0.145 |
0.156 |
| 52 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.144 |
0.293 |
| 53 |
255900_at |
AT1G17830
|
unknown |
0.144 |
0.457 |
| 54 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.143 |
0.079 |
| 55 |
265276_at |
AT2G28400
|
unknown |
0.139 |
-0.073 |
| 56 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
0.137 |
0.302 |
| 57 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
0.136 |
0.406 |
| 58 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.136 |
0.077 |
| 59 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
0.134 |
-0.237 |
| 60 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.129 |
0.344 |
| 61 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.129 |
0.093 |
| 62 |
253246_at |
AT4G34600
|
unknown |
0.127 |
0.265 |
| 63 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.126 |
0.382 |
| 64 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.126 |
0.140 |
| 65 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.126 |
0.054 |
| 66 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.125 |
-0.046 |
| 67 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.124 |
0.478 |
| 68 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.123 |
0.670 |
| 69 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.122 |
-0.064 |
| 70 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.121 |
0.159 |
| 71 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.120 |
0.347 |
| 72 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.117 |
0.145 |
| 73 |
256899_at |
AT3G24660
|
TMKL1, transmembrane kinase-like 1 |
0.117 |
0.121 |
| 74 |
251364_at |
AT3G61300
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.116 |
-0.038 |
| 75 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.114 |
-0.066 |
| 76 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.114 |
0.029 |
| 77 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.113 |
0.147 |
| 78 |
257517_at |
AT3G16330
|
unknown |
0.113 |
0.318 |
| 79 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.112 |
0.086 |
| 80 |
249136_at |
AT5G43180
|
unknown |
0.112 |
0.284 |
| 81 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.110 |
-0.189 |
| 82 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
0.110 |
-0.222 |
| 83 |
251640_at |
AT3G57450
|
unknown |
0.110 |
-0.144 |
| 84 |
251240_at |
AT3G62450
|
unknown |
0.109 |
0.047 |
| 85 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.107 |
0.284 |
| 86 |
249765_at |
AT5G24030
|
SLAH3, SLAC1 homologue 3 |
0.107 |
-0.365 |
| 87 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.105 |
0.035 |
| 88 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.105 |
0.098 |
| 89 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
0.104 |
0.286 |
| 90 |
261455_at |
AT1G21070
|
[Nucleotide-sugar transporter family protein] |
0.104 |
-0.126 |
| 91 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.103 |
-0.172 |
| 92 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
0.103 |
0.200 |
| 93 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.102 |
0.281 |
| 94 |
259030_at |
AT3G09310
|
unknown |
0.102 |
0.159 |
| 95 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.102 |
0.239 |
| 96 |
259864_at |
AT1G72800
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.101 |
0.185 |
| 97 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.101 |
0.092 |
| 98 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.101 |
0.167 |
| 99 |
263840_at |
AT2G36885
|
unknown |
0.101 |
-0.195 |
| 100 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.100 |
0.050 |
| 101 |
256927_at |
AT3G22550
|
unknown |
0.100 |
0.219 |
| 102 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
0.100 |
0.035 |
| 103 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
0.099 |
0.119 |
| 104 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.099 |
0.415 |
| 105 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.098 |
0.467 |
| 106 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.098 |
0.284 |
| 107 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.097 |
0.293 |
| 108 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.096 |
0.165 |
| 109 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.096 |
-0.059 |
| 110 |
258112_at |
AT3G14640
|
CYP72A10, cytochrome P450, family 72, subfamily A, polypeptide 10 |
0.096 |
-0.042 |
| 111 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
0.095 |
0.171 |
| 112 |
255957_at |
AT1G22160
|
unknown |
0.095 |
0.049 |
| 113 |
261545_at |
AT1G63530
|
unknown |
0.095 |
0.081 |
| 114 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.742 |
-0.141 |
| 115 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.467 |
-0.142 |
| 116 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.433 |
-0.135 |
| 117 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.424 |
-0.585 |
| 118 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.380 |
-0.444 |
| 119 |
258468_at |
AT3G06070
|
unknown |
-0.377 |
-1.107 |
| 120 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.353 |
-0.065 |
| 121 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.352 |
-0.042 |
| 122 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.306 |
-0.295 |
| 123 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.288 |
-0.197 |
| 124 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.286 |
-0.479 |
| 125 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.275 |
-0.008 |
| 126 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.267 |
-0.230 |
| 127 |
250327_at |
AT5G12050
|
unknown |
-0.266 |
-0.763 |
| 128 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.261 |
-0.136 |
| 129 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.258 |
-0.636 |
| 130 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.254 |
-0.145 |
| 131 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-0.248 |
0.042 |
| 132 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.245 |
-0.402 |
| 133 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.240 |
-0.052 |
| 134 |
247585_at |
AT5G60680
|
unknown |
-0.240 |
-0.187 |
| 135 |
247754_at |
AT5G59080
|
unknown |
-0.239 |
-0.310 |
| 136 |
250828_at |
AT5G05250
|
unknown |
-0.237 |
0.476 |
| 137 |
261221_at |
AT1G19960
|
unknown |
-0.231 |
-0.251 |
| 138 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.229 |
0.073 |
| 139 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.226 |
-0.329 |
| 140 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.226 |
-0.028 |
| 141 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.219 |
0.076 |
| 142 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.218 |
-0.700 |
| 143 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.217 |
-0.039 |
| 144 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.208 |
0.267 |
| 145 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.207 |
-0.492 |
| 146 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.201 |
-0.631 |
| 147 |
265248_at |
AT2G43010
|
AtPIF4, PIF4, phytochrome interacting factor 4, SRL2 |
-0.201 |
-0.384 |
| 148 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.200 |
-0.414 |
| 149 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-0.199 |
-0.139 |
| 150 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.198 |
0.581 |
| 151 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.197 |
-0.259 |
| 152 |
252427_at |
AT3G47640
|
PYE, POPEYE |
-0.195 |
0.468 |
| 153 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.194 |
-0.166 |
| 154 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.194 |
-0.092 |
| 155 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.192 |
-0.005 |
| 156 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.191 |
0.157 |
| 157 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.190 |
-0.134 |
| 158 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.189 |
-0.092 |
| 159 |
259001_at |
AT3G01960
|
unknown |
-0.188 |
-0.240 |
| 160 |
256225_at |
AT1G56220
|
[Dormancy/auxin associated family protein] |
-0.185 |
-0.062 |
| 161 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.183 |
-0.247 |
| 162 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.182 |
-0.837 |
| 163 |
262170_at |
AT1G74940
|
unknown |
-0.181 |
-0.048 |
| 164 |
247903_at |
AT5G57340
|
unknown |
-0.177 |
0.172 |
| 165 |
265387_at |
AT2G20670
|
unknown |
-0.177 |
-0.374 |
| 166 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.176 |
-0.153 |
| 167 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.174 |
0.047 |
| 168 |
246270_at |
AT4G36500
|
unknown |
-0.168 |
-0.159 |
| 169 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.167 |
0.069 |
| 170 |
265503_at |
AT2G15510
|
[non-LTR retrotransposon family (LINE), has a 3.0e-20 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
-0.164 |
0.003 |
| 171 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.162 |
0.082 |
| 172 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
-0.161 |
-0.049 |
| 173 |
251374_at |
AT3G60390
|
HAT3, homeobox-leucine zipper protein 3 |
-0.160 |
-0.219 |
| 174 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.157 |
0.502 |
| 175 |
247726_at |
AT5G59430
|
ATTRP1, TELOMERE REPEAT BINDING PROTEIN 1, TRP1, telomeric repeat binding protein 1 |
-0.156 |
0.025 |
| 176 |
267344_at |
AT2G44230
|
unknown |
-0.156 |
-0.276 |
| 177 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.154 |
-0.240 |
| 178 |
246997_at |
AT5G67390
|
unknown |
-0.154 |
0.199 |
| 179 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.153 |
-0.027 |
| 180 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.152 |
-0.518 |
| 181 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.152 |
-0.152 |
| 182 |
264314_at |
AT1G70420
|
unknown |
-0.150 |
0.208 |
| 183 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.147 |
-0.123 |
| 184 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.146 |
-0.046 |
| 185 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
-0.146 |
-0.213 |
| 186 |
247029_at |
AT5G67190
|
DEAR2, DREB and EAR motif protein 2 |
-0.145 |
-0.284 |
| 187 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-0.142 |
0.169 |
| 188 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.142 |
-0.341 |
| 189 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.142 |
0.223 |
| 190 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.142 |
-0.174 |
| 191 |
245906_at |
AT5G11070
|
unknown |
-0.142 |
-0.126 |
| 192 |
249941_at |
AT5G22270
|
unknown |
-0.141 |
-0.093 |
| 193 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
-0.141 |
0.387 |
| 194 |
252520_at |
AT3G46370
|
[Leucine-rich repeat protein kinase family protein] |
-0.140 |
-0.130 |
| 195 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.140 |
-0.042 |
| 196 |
262399_at |
AT1G49500
|
unknown |
-0.140 |
-0.061 |
| 197 |
264379_at |
AT2G25200
|
unknown |
-0.139 |
-0.355 |
| 198 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.139 |
0.059 |
| 199 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.139 |
-0.410 |
| 200 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
-0.139 |
0.016 |
| 201 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.138 |
-0.146 |
| 202 |
250734_at |
AT5G06270
|
unknown |
-0.137 |
-0.088 |
| 203 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.136 |
-0.144 |
| 204 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
-0.134 |
-0.250 |
| 205 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.134 |
0.057 |
| 206 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
-0.134 |
-0.150 |
| 207 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.133 |
-0.610 |
| 208 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.133 |
-0.235 |
| 209 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.132 |
-0.274 |
| 210 |
248801_at |
AT5G47370
|
HAT2 |
-0.132 |
-0.378 |
| 211 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
-0.132 |
0.275 |
| 212 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.132 |
-0.376 |
| 213 |
259373_at |
AT1G69160
|
unknown |
-0.130 |
-0.194 |
| 214 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
-0.130 |
0.030 |
| 215 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.129 |
-0.094 |
| 216 |
252425_at |
AT3G47620
|
AtTCP14, TEOSINTE BRANCHED, cycloidea and PCF (TCP) 14, TCP14, TEOSINTE BRANCHED, cycloidea and PCF (TCP) 14 |
-0.129 |
-0.192 |
| 217 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.128 |
0.344 |
| 218 |
263305_at |
AT2G01930
|
ATBPC1, BASIC PENTACYSTEINE1, BBR, BPC1, basic pentacysteine1 |
-0.128 |
-0.174 |
| 219 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.128 |
-0.225 |
| 220 |
260303_at |
AT1G70520
|
ASG6, ALTERED SEED GERMINATION 6, CRK2, cysteine-rich RLK (RECEPTOR-like protein kinase) 2 |
-0.127 |
-0.199 |
| 221 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.127 |
-0.009 |
| 222 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.127 |
-0.180 |
| 223 |
261772_at |
AT1G76240
|
unknown |
-0.127 |
-0.142 |
| 224 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.126 |
-0.032 |
| 225 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.126 |
0.141 |