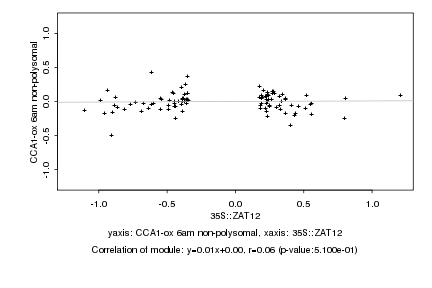
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
35S::ZAT12 |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
1.205 |
0.089 |
| 2 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.802 |
0.054 |
| 3 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.797 |
-0.237 |
| 4 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.553 |
-0.028 |
| 5 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
0.552 |
-0.184 |
| 6 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.546 |
-0.033 |
| 7 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.517 |
0.098 |
| 8 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.509 |
-0.096 |
| 9 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.461 |
-0.071 |
| 10 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
0.438 |
-0.170 |
| 11 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.425 |
-0.191 |
| 12 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.406 |
-0.048 |
| 13 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.399 |
-0.344 |
| 14 |
251050_at |
AT5G02440
|
unknown |
0.363 |
0.035 |
| 15 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.362 |
-0.164 |
| 16 |
262801_at |
AT1G21010
|
unknown |
0.360 |
0.045 |
| 17 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.340 |
0.108 |
| 18 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.331 |
0.006 |
| 19 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
0.323 |
-0.107 |
| 20 |
247126_at |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
0.320 |
0.083 |
| 21 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
0.318 |
-0.045 |
| 22 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.300 |
-0.076 |
| 23 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.284 |
0.119 |
| 24 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.272 |
0.160 |
| 25 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.265 |
0.122 |
| 26 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.264 |
0.151 |
| 27 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.258 |
0.030 |
| 28 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.246 |
-0.056 |
| 29 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.243 |
-0.062 |
| 30 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.240 |
0.030 |
| 31 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.238 |
0.093 |
| 32 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.234 |
0.011 |
| 33 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.234 |
0.147 |
| 34 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
0.231 |
0.114 |
| 35 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.230 |
-0.210 |
| 36 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
0.226 |
0.079 |
| 37 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.224 |
0.033 |
| 38 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.223 |
-0.133 |
| 39 |
260771_at |
AT1G49160
|
WNK7 |
0.221 |
-0.027 |
| 40 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.218 |
0.076 |
| 41 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.214 |
-0.088 |
| 42 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.201 |
0.176 |
| 43 |
264880_at |
AT1G61210
|
DWA3, DWD hypersensitive to ABA 3 |
0.197 |
0.072 |
| 44 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.188 |
-0.029 |
| 45 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.186 |
0.094 |
| 46 |
250753_at |
AT5G05860
|
UGT76C2, UDP-glucosyl transferase 76C2 |
0.186 |
0.046 |
| 47 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
0.182 |
-0.055 |
| 48 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
0.181 |
-0.092 |
| 49 |
259912_at |
AT1G72670
|
iqd8, IQ-domain 8 |
0.175 |
0.062 |
| 50 |
256756_at |
AT3G25610
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.175 |
0.227 |
| 51 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-1.106 |
-0.128 |
| 52 |
261221_at |
AT1G19960
|
unknown |
-0.988 |
0.019 |
| 53 |
245906_at |
AT5G11070
|
unknown |
-0.959 |
-0.175 |
| 54 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.938 |
0.162 |
| 55 |
256603_at |
AT3G28270
|
unknown |
-0.907 |
-0.496 |
| 56 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.901 |
-0.157 |
| 57 |
260012_at |
AT1G67865
|
unknown |
-0.892 |
-0.049 |
| 58 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.879 |
0.064 |
| 59 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.870 |
-0.079 |
| 60 |
254574_at |
AT4G19430
|
unknown |
-0.816 |
-0.110 |
| 61 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.768 |
-0.040 |
| 62 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.737 |
-0.009 |
| 63 |
260495_at |
AT2G41810
|
unknown |
-0.688 |
-0.144 |
| 64 |
267459_at |
AT2G33850
|
unknown |
-0.678 |
-0.016 |
| 65 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.637 |
-0.088 |
| 66 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.621 |
0.437 |
| 67 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.617 |
-0.036 |
| 68 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.604 |
-0.024 |
| 69 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.550 |
0.058 |
| 70 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.549 |
-0.106 |
| 71 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.544 |
0.038 |
| 72 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
-0.494 |
-0.115 |
| 73 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.490 |
-0.052 |
| 74 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.488 |
0.016 |
| 75 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
-0.462 |
0.133 |
| 76 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.456 |
0.122 |
| 77 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.450 |
0.003 |
| 78 |
258879_at |
AT3G03270
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.449 |
-0.027 |
| 79 |
252983_at |
AT4G37980
|
ATCAD7, CAD7, CINNAMYL-ALCOHOL DEHYDROGENASE 7, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-1, elicitor-activated gene 3-1 |
-0.448 |
-0.060 |
| 80 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.440 |
-0.249 |
| 81 |
265768_at |
AT2G48020
|
[Major facilitator superfamily protein] |
-0.439 |
-0.073 |
| 82 |
247964_at |
AT5G56600
|
PFN3, PROFILIN 3, PRF3, profilin 3 |
-0.424 |
0.013 |
| 83 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.402 |
-0.030 |
| 84 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.399 |
0.210 |
| 85 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
-0.394 |
-0.136 |
| 86 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.390 |
0.040 |
| 87 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.389 |
0.049 |
| 88 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.388 |
0.005 |
| 89 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.381 |
0.056 |
| 90 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.377 |
0.010 |
| 91 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.376 |
0.115 |
| 92 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.367 |
0.260 |
| 93 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.362 |
0.032 |
| 94 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.362 |
-0.019 |
| 95 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.358 |
0.380 |
| 96 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.357 |
0.018 |
| 97 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.355 |
0.041 |
| 98 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.354 |
0.126 |
| 99 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.354 |
0.121 |
| 100 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
-0.350 |
0.022 |