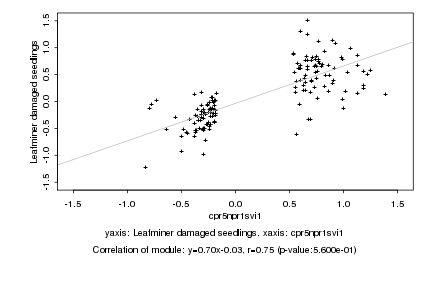
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cpr5npr1svi1 |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
1.384 |
0.141 |
| 2 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
1.244 |
0.580 |
| 3 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.214 |
0.514 |
| 4 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.184 |
0.300 |
| 5 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
1.178 |
0.568 |
| 6 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
1.177 |
0.257 |
| 7 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.129 |
0.162 |
| 8 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
1.129 |
0.675 |
| 9 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
1.122 |
0.856 |
| 10 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
1.059 |
0.987 |
| 11 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
1.030 |
0.545 |
| 12 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
1.014 |
0.199 |
| 13 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
1.000 |
-0.128 |
| 14 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.983 |
0.795 |
| 15 |
261545_at |
AT1G63530
|
unknown |
0.982 |
0.051 |
| 16 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.977 |
0.830 |
| 17 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.925 |
1.085 |
| 18 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.913 |
0.621 |
| 19 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.904 |
0.395 |
| 20 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.895 |
1.150 |
| 21 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.895 |
0.344 |
| 22 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.864 |
0.487 |
| 23 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.861 |
0.685 |
| 24 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.861 |
0.197 |
| 25 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.832 |
0.501 |
| 26 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.824 |
0.939 |
| 27 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
0.822 |
0.289 |
| 28 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.804 |
0.701 |
| 29 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.791 |
0.669 |
| 30 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.769 |
0.715 |
| 31 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.767 |
0.748 |
| 32 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.767 |
0.783 |
| 33 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.761 |
1.122 |
| 34 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.758 |
0.057 |
| 35 |
266017_at |
AT2G18690
|
unknown |
0.757 |
0.560 |
| 36 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.744 |
0.655 |
| 37 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.743 |
0.841 |
| 38 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.742 |
0.442 |
| 39 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.740 |
0.674 |
| 40 |
267178_at |
AT2G37750
|
unknown |
0.739 |
0.551 |
| 41 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.734 |
0.786 |
| 42 |
258203_at |
AT3G13950
|
unknown |
0.731 |
0.665 |
| 43 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.726 |
0.275 |
| 44 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.707 |
0.828 |
| 45 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.702 |
0.778 |
| 46 |
264635_at |
AT1G65500
|
unknown |
0.696 |
0.405 |
| 47 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.696 |
0.376 |
| 48 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.693 |
0.181 |
| 49 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.686 |
-0.334 |
| 50 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.670 |
-0.321 |
| 51 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.665 |
1.262 |
| 52 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.662 |
0.658 |
| 53 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
0.662 |
0.773 |
| 54 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.661 |
1.524 |
| 55 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.660 |
0.609 |
| 56 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.650 |
0.839 |
| 57 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.648 |
0.207 |
| 58 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.647 |
0.780 |
| 59 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.646 |
0.496 |
| 60 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.642 |
0.371 |
| 61 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.633 |
0.436 |
| 62 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.627 |
0.311 |
| 63 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.623 |
0.215 |
| 64 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.598 |
1.304 |
| 65 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.597 |
0.622 |
| 66 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.596 |
0.394 |
| 67 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.594 |
0.685 |
| 68 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.591 |
0.629 |
| 69 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.587 |
-0.040 |
| 70 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
0.575 |
0.626 |
| 71 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.569 |
0.716 |
| 72 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.561 |
0.384 |
| 73 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.561 |
-0.611 |
| 74 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.554 |
0.172 |
| 75 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.547 |
0.267 |
| 76 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.543 |
0.543 |
| 77 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.536 |
0.902 |
| 78 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.534 |
0.882 |
| 79 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.833 |
-1.219 |
| 80 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.798 |
-0.121 |
| 81 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.783 |
-0.038 |
| 82 |
262346_at |
AT1G63980
|
[D111/G-patch domain-containing protein] |
-0.735 |
0.025 |
| 83 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.644 |
-0.505 |
| 84 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.555 |
-0.279 |
| 85 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.506 |
-0.918 |
| 86 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.503 |
-0.650 |
| 87 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.487 |
-0.514 |
| 88 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.455 |
-0.559 |
| 89 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.444 |
-0.586 |
| 90 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.426 |
-0.319 |
| 91 |
247486_at |
AT5G62140
|
unknown |
-0.386 |
-0.407 |
| 92 |
253814_at |
AT4G28290
|
unknown |
-0.381 |
0.149 |
| 93 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.379 |
-0.636 |
| 94 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.375 |
-0.526 |
| 95 |
265387_at |
AT2G20670
|
unknown |
-0.374 |
-0.242 |
| 96 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.371 |
-0.566 |
| 97 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.364 |
-0.525 |
| 98 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.358 |
-0.271 |
| 99 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.352 |
-0.136 |
| 100 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
-0.348 |
-0.348 |
| 101 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.346 |
-0.348 |
| 102 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.334 |
-0.496 |
| 103 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
-0.326 |
-0.349 |
| 104 |
245724_at |
AT1G73390
|
[Endosomal targeting BRO1-like domain-containing protein] |
-0.323 |
0.174 |
| 105 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.321 |
-0.071 |
| 106 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.320 |
-0.286 |
| 107 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.319 |
-0.231 |
| 108 |
258315_at |
AT3G16175
|
[Thioesterase superfamily protein] |
-0.317 |
-0.237 |
| 109 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.315 |
-0.168 |
| 110 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.312 |
-0.503 |
| 111 |
258782_at |
AT3G11750
|
FOLB1 |
-0.304 |
-0.315 |
| 112 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
-0.302 |
-0.141 |
| 113 |
252011_at |
AT3G52720
|
ACA1, alpha carbonic anhydrase 1, ATACA1, A. THALIANA ALPHA CARBONIC ANHYDRASE 1, CAH1, CARBONIC ANHYDRASE 1 |
-0.301 |
-0.984 |
| 114 |
264909_at |
AT2G17300
|
unknown |
-0.301 |
-0.537 |
| 115 |
256892_at |
AT3G19000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.299 |
-0.228 |
| 116 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.298 |
-0.496 |
| 117 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.292 |
-0.486 |
| 118 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
-0.292 |
-0.193 |
| 119 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.280 |
-0.707 |
| 120 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.278 |
-0.239 |
| 121 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.272 |
-0.420 |
| 122 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
-0.267 |
-0.059 |
| 123 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.265 |
-0.422 |
| 124 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.262 |
-0.048 |
| 125 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.258 |
-0.195 |
| 126 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
-0.257 |
-0.388 |
| 127 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
-0.247 |
-0.005 |
| 128 |
251142_at |
AT5G01015
|
unknown |
-0.247 |
-0.462 |
| 129 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.246 |
-0.142 |
| 130 |
261350_at |
AT1G79770
|
unknown |
-0.246 |
-0.510 |
| 131 |
253478_at |
AT4G32350
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.234 |
-0.270 |
| 132 |
250413_at |
AT5G11160
|
APT5, adenine phosphoribosyltransferase 5 |
-0.231 |
-0.189 |
| 133 |
249288_at |
AT5G41050
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.231 |
-0.410 |
| 134 |
264378_at |
AT2G25220
|
[Protein kinase superfamily protein] |
-0.231 |
-0.123 |
| 135 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.227 |
0.086 |
| 136 |
264331_at |
AT1G04130
|
AtTPR2, TPR2, tetratricopeptide repeat 2 |
-0.223 |
-0.131 |
| 137 |
260491_at |
AT1G51440
|
DALL2, DAD1-Like Lipase 2 |
-0.221 |
0.020 |
| 138 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.218 |
-0.124 |
| 139 |
246915_at |
AT5G25880
|
ATNADP-ME3, Arabidopsis thaliana NADP-malic enzyme 3, NADP-ME3, NADP-malic enzyme 3 |
-0.216 |
0.092 |
| 140 |
250074_at |
AT5G17310
|
AtUGP2, UDP-GLUCOSE PYROPHOSPHORYLASE 2, UGP2, UDP-glucose pyrophosphorylase 2 |
-0.215 |
-0.261 |
| 141 |
245984_at |
AT5G13090
|
unknown |
-0.212 |
-0.219 |
| 142 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.207 |
-0.007 |
| 143 |
256985_at |
AT3G13540
|
ATMYB5, myb domain protein 5, MYB5, myb domain protein 5 |
-0.203 |
0.009 |
| 144 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.203 |
-0.386 |
| 145 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
-0.198 |
-0.143 |
| 146 |
251605_at |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
-0.197 |
-0.378 |
| 147 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.195 |
-0.361 |
| 148 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.194 |
-0.060 |
| 149 |
260875_at |
AT1G21410
|
SKP2A |
-0.194 |
0.030 |
| 150 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.191 |
0.032 |
| 151 |
251150_at |
AT3G63120
|
CYCP1;1, cyclin p1;1 |
-0.190 |
-0.220 |
| 152 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.187 |
-0.154 |
| 153 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.187 |
-0.251 |
| 154 |
248336_at |
AT5G52420
|
unknown |
-0.185 |
-0.208 |
| 155 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.183 |
0.150 |