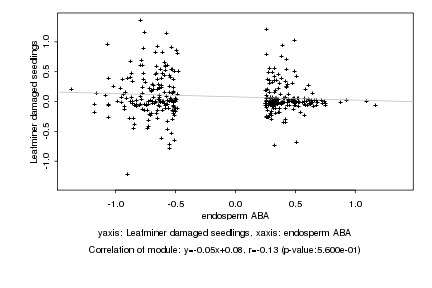
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
endosperm ABA |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.167 |
-0.052 |
| 2 |
267321_at |
AT2G19320
|
unknown |
1.087 |
0.002 |
| 3 |
252914_at |
AT4G39130
|
[Dehydrin family protein] |
0.924 |
0.032 |
| 4 |
250118_at |
AT5G16460
|
[Putative adipose-regulatory protein (Seipin)] |
0.869 |
-0.015 |
| 5 |
253494_at |
AT4G31830
|
unknown |
0.750 |
-0.020 |
| 6 |
255049_at |
AT4G09610
|
GASA2, GAST1 protein homolog 2 |
0.747 |
-0.051 |
| 7 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
0.737 |
0.016 |
| 8 |
262734_at |
AT1G28640
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.720 |
-0.023 |
| 9 |
248969_at |
AT5G45310
|
unknown |
0.718 |
0.028 |
| 10 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
0.679 |
-0.004 |
| 11 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.675 |
-0.053 |
| 12 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
0.670 |
0.018 |
| 13 |
247096_at |
AT5G66430
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.658 |
0.023 |
| 14 |
260458_at |
AT1G68250
|
unknown |
0.651 |
-0.067 |
| 15 |
267036_at |
AT2G38465
|
unknown |
0.641 |
0.138 |
| 16 |
255840_at |
AT2G33520
|
unknown |
0.639 |
-0.007 |
| 17 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.627 |
0.029 |
| 18 |
255493_at |
AT4G02690
|
AtLFG3, LFG3, LIFEGUARD 3 |
0.623 |
-0.019 |
| 19 |
265714_at |
AT2G03520
|
ATUPS4, ureide permease 4, UPS4, ureide permease 4 |
0.619 |
-0.044 |
| 20 |
248096_at |
AT5G55240
|
ATPXG2, ARABIDOPSIS THALIANA PEROXYGENASE 2 |
0.615 |
0.027 |
| 21 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
0.611 |
-0.017 |
| 22 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.609 |
0.035 |
| 23 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.602 |
0.285 |
| 24 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.598 |
-0.053 |
| 25 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.587 |
0.010 |
| 26 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.584 |
0.213 |
| 27 |
262069_at |
AT1G80090
|
CBSX4, CBS domain containing protein 4 |
0.584 |
-0.062 |
| 28 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.580 |
-0.058 |
| 29 |
257059_at |
AT3G15280
|
unknown |
0.572 |
0.060 |
| 30 |
251731_at |
AT3G56350
|
[Iron/manganese superoxide dismutase family protein] |
0.571 |
-0.047 |
| 31 |
259615_at |
AT1G47980
|
unknown |
0.568 |
0.012 |
| 32 |
255763_at |
AT1G16730
|
unknown |
0.568 |
-0.219 |
| 33 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.559 |
-0.016 |
| 34 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.542 |
-0.181 |
| 35 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.531 |
-0.006 |
| 36 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
0.526 |
-0.075 |
| 37 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
0.524 |
0.015 |
| 38 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.522 |
0.057 |
| 39 |
258873_at |
AT3G03240
|
[alpha/beta-Hydrolases superfamily protein] |
0.518 |
-0.013 |
| 40 |
253322_at |
AT4G33980
|
unknown |
0.517 |
0.060 |
| 41 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.508 |
0.427 |
| 42 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.504 |
-0.009 |
| 43 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.503 |
-0.678 |
| 44 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
0.494 |
0.016 |
| 45 |
267313_at |
AT2G34740
|
[Protein phosphatase 2C family protein] |
0.493 |
-0.062 |
| 46 |
248076_at |
AT5G55750
|
[hydroxyproline-rich glycoprotein family protein] |
0.491 |
-0.025 |
| 47 |
259782_at |
AT1G29680
|
unknown |
0.491 |
0.053 |
| 48 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.487 |
0.019 |
| 49 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.487 |
0.509 |
| 50 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.487 |
-0.061 |
| 51 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.487 |
1.035 |
| 52 |
245554_at |
AT4G15380
|
CYP705A4, cytochrome P450, family 705, subfamily A, polypeptide 4 |
0.484 |
0.002 |
| 53 |
253152_at |
AT4G35690
|
unknown |
0.483 |
-0.004 |
| 54 |
247861_at |
AT5G58160
|
[actin binding] |
0.477 |
0.054 |
| 55 |
265828_at |
AT2G14520
|
unknown |
0.477 |
-0.131 |
| 56 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
0.475 |
0.043 |
| 57 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.474 |
0.015 |
| 58 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.473 |
0.307 |
| 59 |
267579_at |
AT2G42000
|
AtMT4a, Arabidopsis thaliana metallothionein 4a |
0.460 |
0.013 |
| 60 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.451 |
0.002 |
| 61 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.449 |
-0.059 |
| 62 |
259746_at |
AT1G71060
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.440 |
0.049 |
| 63 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
0.439 |
0.028 |
| 64 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.436 |
0.119 |
| 65 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.431 |
0.288 |
| 66 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
0.430 |
-0.063 |
| 67 |
246582_at |
AT1G31750
|
[proline-rich family protein] |
0.427 |
-0.007 |
| 68 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.424 |
0.261 |
| 69 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.423 |
0.716 |
| 70 |
250826_at |
AT5G05220
|
unknown |
0.422 |
0.239 |
| 71 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.420 |
0.553 |
| 72 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.416 |
-0.297 |
| 73 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.416 |
0.082 |
| 74 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.415 |
0.336 |
| 75 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.415 |
-0.108 |
| 76 |
264352_at |
AT1G03270
|
unknown |
0.413 |
-0.343 |
| 77 |
248428_at |
AT5G51760
|
AHG1, ABA-hypersensitive germination 1 |
0.409 |
0.066 |
| 78 |
247172_at |
AT5G65550
|
[UDP-Glycosyltransferase superfamily protein] |
0.408 |
-0.009 |
| 79 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.401 |
-0.339 |
| 80 |
247535_at |
AT5G61620
|
[myb-like transcription factor family protein] |
0.399 |
0.066 |
| 81 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.394 |
-0.015 |
| 82 |
255761_at |
AT1G16770
|
unknown |
0.391 |
-0.013 |
| 83 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.388 |
0.951 |
| 84 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
0.385 |
-0.013 |
| 85 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.383 |
-0.047 |
| 86 |
260005_at |
AT1G67920
|
unknown |
0.382 |
0.768 |
| 87 |
246332_at |
AT3G44830
|
[Lecithin:cholesterol acyltransferase family protein] |
0.381 |
-0.008 |
| 88 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.380 |
0.393 |
| 89 |
265095_at |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
0.374 |
-0.024 |
| 90 |
256416_at |
AT3G11050
|
ATFER2, ferritin 2, FER2, ferritin 2 |
0.374 |
0.120 |
| 91 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.366 |
-0.102 |
| 92 |
248614_at |
AT5G49560
|
[Putative methyltransferase family protein] |
0.360 |
-0.168 |
| 93 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.360 |
0.308 |
| 94 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.356 |
-0.170 |
| 95 |
258264_at |
AT3G15790
|
ATMBD11, MBD11, methyl-CPG-binding domain 11 |
0.355 |
0.220 |
| 96 |
262416_at |
AT1G49390
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.354 |
0.047 |
| 97 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.354 |
-0.010 |
| 98 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.352 |
0.469 |
| 99 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.348 |
-0.031 |
| 100 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.345 |
-0.035 |
| 101 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.344 |
0.038 |
| 102 |
253019_at |
AT4G38010
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.339 |
-0.026 |
| 103 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.337 |
0.367 |
| 104 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.336 |
0.062 |
| 105 |
253154_at |
AT4G35710
|
unknown |
0.336 |
0.030 |
| 106 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
0.335 |
-0.024 |
| 107 |
262185_at |
AT1G77950
|
AGL67, AGAMOUS-like 67 |
0.335 |
-0.022 |
| 108 |
246016_at |
AT5G10720
|
AHK5, histidine kinase 5, CKI2, CYTOKININ INDEPENDENT 2, HK5, histidine kinase 5 |
0.334 |
0.053 |
| 109 |
266948_at |
AT2G18850
|
[SET domain-containing protein] |
0.334 |
-0.189 |
| 110 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.334 |
0.192 |
| 111 |
248084_at |
AT5G55470
|
ATNHX3, ARABIDOPSIS THALIANA NA+/H+ (SODIUM HYDROGEN) EXCHANGER 3, NHX3, Na+/H+ (sodium hydrogen) exchanger 3 |
0.333 |
-0.109 |
| 112 |
260498_at |
AT2G41710
|
[Integrase-type DNA-binding superfamily protein] |
0.331 |
-0.021 |
| 113 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.330 |
-0.039 |
| 114 |
247634_at |
AT5G60520
|
[Late embryogenesis abundant (LEA) protein-related] |
0.330 |
-0.022 |
| 115 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.326 |
0.003 |
| 116 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.321 |
0.360 |
| 117 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
0.321 |
-0.059 |
| 118 |
254201_at |
AT4G24130
|
unknown |
0.320 |
0.154 |
| 119 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
0.320 |
-0.721 |
| 120 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.320 |
0.430 |
| 121 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.319 |
0.565 |
| 122 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.318 |
0.040 |
| 123 |
258254_at |
AT3G26782
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.317 |
-0.028 |
| 124 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.315 |
-0.016 |
| 125 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.313 |
-0.002 |
| 126 |
251383_at |
AT3G60740
|
CHO, CHAMPIGNON, EMB133, EMBRYO DEFECTIVE 133, TFC D, TUBULIN FOLDING COFACTOR D, TTN1, TITAN 1 |
0.313 |
-0.110 |
| 127 |
262201_at |
AT2G01120
|
ATORC4, ORIGIN RECOGNITION COMPLEX SUBUNIT 4, ORC4, origin recognition complex subunit 4 |
0.312 |
-0.073 |
| 128 |
267445_at |
AT2G33680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.312 |
-0.067 |
| 129 |
253969_at |
AT4G26430
|
CSN6B, COP9 signalosome subunit 6B |
0.309 |
-0.072 |
| 130 |
263138_at |
AT1G65090
|
unknown |
0.309 |
0.019 |
| 131 |
246383_at |
AT1G77360
|
APPR6, Arabidopsis pentatricopeptide repeat 6 |
0.304 |
0.322 |
| 132 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.304 |
0.071 |
| 133 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.301 |
0.495 |
| 134 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
0.297 |
-0.301 |
| 135 |
248915_at |
AT5G45690
|
unknown |
0.297 |
0.014 |
| 136 |
266733_at |
AT2G03280
|
[O-fucosyltransferase family protein] |
0.294 |
-0.160 |
| 137 |
251202_at |
AT3G63040
|
unknown |
0.294 |
-0.043 |
| 138 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
0.293 |
-0.224 |
| 139 |
260598_at |
AT1G55930
|
[CBS domain-containing protein / transporter associated domain-containing protein] |
0.292 |
-0.199 |
| 140 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
0.291 |
-0.007 |
| 141 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.291 |
0.071 |
| 142 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.291 |
0.084 |
| 143 |
255300_at |
AT4G04870
|
CLS, cardiolipin synthase |
0.290 |
-0.051 |
| 144 |
259586_at |
AT1G28100
|
unknown |
0.287 |
-0.055 |
| 145 |
258845_at |
AT3G03150
|
unknown |
0.287 |
0.061 |
| 146 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.285 |
0.054 |
| 147 |
262010_at |
AT1G35612
|
[expressed protein] |
0.285 |
-0.027 |
| 148 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.284 |
-0.223 |
| 149 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.281 |
0.314 |
| 150 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
0.281 |
0.364 |
| 151 |
250874_at |
AT5G04010
|
[F-box family protein] |
0.279 |
-0.040 |
| 152 |
260897_at |
AT1G29330
|
AERD2, ARABIDOPSIS ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ATERD2, ARABIDOPSIS THALIANA ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2, ERD2, ENDOPLASMIC RETICULUM RETENTION DEFECTIVE 2 |
0.277 |
0.570 |
| 153 |
266490_at |
AT2G07000
|
unknown |
0.277 |
0.007 |
| 154 |
249409_at |
AT5G40340
|
[Tudor/PWWP/MBT superfamily protein] |
0.276 |
0.165 |
| 155 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
0.275 |
-0.060 |
| 156 |
246973_at |
AT5G24970
|
[Protein kinase superfamily protein] |
0.274 |
-0.263 |
| 157 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.272 |
0.490 |
| 158 |
257937_at |
AT3G19810
|
unknown |
0.270 |
-0.020 |
| 159 |
252468_at |
AT3G46970
|
ATPHS2, Arabidopsis thaliana alpha-glucan phosphorylase 2, PHS2, alpha-glucan phosphorylase 2 |
0.265 |
0.084 |
| 160 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.264 |
-0.144 |
| 161 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.262 |
0.382 |
| 162 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.261 |
0.207 |
| 163 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
0.260 |
-0.253 |
| 164 |
245508_at |
AT4G15720
|
REME2, Required for Efficiency of Mitochondrial Editing 2 |
0.259 |
0.030 |
| 165 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.258 |
0.803 |
| 166 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
0.258 |
0.193 |
| 167 |
246386_at |
AT1G77410
|
BGAL16, beta-galactosidase 16 |
0.257 |
-0.030 |
| 168 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
0.256 |
0.063 |
| 169 |
260641_at |
AT1G53200
|
unknown |
0.254 |
-0.095 |
| 170 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.254 |
-0.066 |
| 171 |
249420_at |
AT5G39820
|
anac094, NAC domain containing protein 94, NAC094, NAC domain containing protein 94 |
0.254 |
0.017 |
| 172 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.253 |
-0.247 |
| 173 |
255961_at |
AT1G22340
|
AtUGT85A7, UDP-glucosyl transferase 85A7, UGT85A7, UDP-glucosyl transferase 85A7 |
0.253 |
0.102 |
| 174 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.252 |
0.005 |
| 175 |
250074_at |
AT5G17310
|
AtUGP2, UDP-GLUCOSE PYROPHOSPHORYLASE 2, UGP2, UDP-glucose pyrophosphorylase 2 |
0.251 |
-0.261 |
| 176 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.251 |
1.222 |
| 177 |
258211_at |
AT3G17890
|
unknown |
0.250 |
-0.065 |
| 178 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.250 |
0.200 |
| 179 |
264241_at |
AT1G54840
|
[HSP20-like chaperones superfamily protein] |
0.250 |
-0.029 |
| 180 |
257841_at |
AT3G25260
|
AtNPF4.1, NPF4.1, NRT1/ PTR family 4.1 |
0.249 |
-0.023 |
| 181 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-1.374 |
0.212 |
| 182 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-1.182 |
-0.177 |
| 183 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
-1.179 |
-0.046 |
| 184 |
253814_at |
AT4G28290
|
unknown |
-1.166 |
0.149 |
| 185 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.086 |
0.105 |
| 186 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-1.070 |
0.965 |
| 187 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
-1.067 |
-0.061 |
| 188 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-1.066 |
-0.044 |
| 189 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-1.066 |
-0.255 |
| 190 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-1.064 |
0.393 |
| 191 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-1.020 |
0.267 |
| 192 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.985 |
0.014 |
| 193 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.966 |
0.229 |
| 194 |
266562_at |
AT2G23970
|
[Class I glutamine amidotransferase-like superfamily protein] |
-0.958 |
-0.009 |
| 195 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.953 |
0.068 |
| 196 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.950 |
0.373 |
| 197 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.937 |
0.125 |
| 198 |
248824_at |
AT5G46940
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.934 |
-0.072 |
| 199 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.930 |
-0.117 |
| 200 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.925 |
0.155 |
| 201 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.910 |
0.058 |
| 202 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.908 |
0.399 |
| 203 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.901 |
-1.219 |
| 204 |
258468_at |
AT3G06070
|
unknown |
-0.885 |
-0.278 |
| 205 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.880 |
0.413 |
| 206 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.879 |
0.012 |
| 207 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.878 |
0.672 |
| 208 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
-0.873 |
0.476 |
| 209 |
253934_at |
AT4G26830
|
[O-Glycosyl hydrolases family 17 protein] |
-0.869 |
0.070 |
| 210 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
-0.867 |
-0.021 |
| 211 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
-0.867 |
0.350 |
| 212 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.864 |
-0.020 |
| 213 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.864 |
0.016 |
| 214 |
263121_at |
AT1G78530
|
[Protein kinase superfamily protein] |
-0.860 |
-0.020 |
| 215 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.854 |
-0.268 |
| 216 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.851 |
-0.437 |
| 217 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.847 |
-0.065 |
| 218 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.844 |
-0.381 |
| 219 |
252347_at |
AT3G48130
|
RSU1 |
-0.833 |
0.022 |
| 220 |
260265_at |
AT1G68510
|
LBD42, LOB domain-containing protein 42 |
-0.832 |
-0.038 |
| 221 |
250535_at |
AT5G08480
|
[VQ motif-containing protein] |
-0.832 |
-0.071 |
| 222 |
245705_at |
AT5G04390
|
[C2H2-type zinc finger family protein] |
-0.805 |
-0.057 |
| 223 |
257490_x_at |
AT1G01380
|
ETC1, ENHANCER OF TRY AND CPC 1 |
-0.797 |
-0.049 |
| 224 |
259286_at |
AT3G11480
|
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
-0.794 |
1.360 |
| 225 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.793 |
0.087 |
| 226 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.793 |
0.614 |
| 227 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.792 |
0.697 |
| 228 |
261878_at |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-0.786 |
0.039 |
| 229 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.780 |
0.610 |
| 230 |
263363_at |
AT2G03850
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.780 |
0.142 |
| 231 |
250772_at |
AT5G05420
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.779 |
-0.125 |
| 232 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.776 |
0.247 |
| 233 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
-0.775 |
0.481 |
| 234 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.772 |
0.370 |
| 235 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.771 |
0.894 |
| 236 |
259996_at |
AT1G67910
|
unknown |
-0.764 |
-0.150 |
| 237 |
263182_at |
AT1G05575
|
unknown |
-0.760 |
1.162 |
| 238 |
254301_at |
AT4G22790
|
[MATE efflux family protein] |
-0.759 |
-0.034 |
| 239 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.753 |
0.325 |
| 240 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
-0.747 |
-0.053 |
| 241 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
-0.747 |
-0.046 |
| 242 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.740 |
-0.438 |
| 243 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.735 |
-0.138 |
| 244 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.731 |
-0.316 |
| 245 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.727 |
0.005 |
| 246 |
258571_at |
AT3G04420
|
anac048, NAC domain containing protein 48, NAC048, NAC domain containing protein 48 |
-0.726 |
-0.045 |
| 247 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.726 |
-0.412 |
| 248 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.725 |
0.224 |
| 249 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
-0.724 |
-0.216 |
| 250 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.716 |
0.028 |
| 251 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.710 |
-0.228 |
| 252 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.708 |
0.171 |
| 253 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.707 |
-0.200 |
| 254 |
265720_at |
AT2G40110
|
[Yippee family putative zinc-binding protein] |
-0.703 |
0.186 |
| 255 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
-0.699 |
-0.027 |
| 256 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
-0.694 |
0.294 |
| 257 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-0.693 |
0.006 |
| 258 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.692 |
0.278 |
| 259 |
259134_at |
AT3G05390
|
unknown |
-0.692 |
0.202 |
| 260 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.686 |
0.074 |
| 261 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.676 |
-0.020 |
| 262 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.675 |
0.002 |
| 263 |
261466_at |
AT1G07690
|
unknown |
-0.673 |
-0.006 |
| 264 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
-0.673 |
0.837 |
| 265 |
260656_at |
AT1G19380
|
unknown |
-0.672 |
0.457 |
| 266 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.666 |
0.591 |
| 267 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
-0.662 |
0.484 |
| 268 |
248443_at |
AT5G51310
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.659 |
-0.194 |
| 269 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
-0.657 |
0.370 |
| 270 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
-0.656 |
0.935 |
| 271 |
264956_at |
AT1G76990
|
ACR3, ACT domain repeat 3 |
-0.654 |
-0.275 |
| 272 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.653 |
-0.139 |
| 273 |
262935_at |
AT1G79410
|
AtOCT5, organic cation/carnitine transporter5, OCT5, organic cation/carnitine transporter5 |
-0.652 |
0.205 |
| 274 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
-0.650 |
0.291 |
| 275 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.648 |
0.046 |
| 276 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
-0.648 |
-0.065 |
| 277 |
254292_at |
AT4G23030
|
[MATE efflux family protein] |
-0.640 |
0.234 |
| 278 |
255860_at |
AT5G34940
|
AtGUS3, glucuronidase 3, GUS3, glucuronidase 3 |
-0.638 |
0.074 |
| 279 |
248568_at |
AT5G49760
|
[Leucine-rich repeat protein kinase family protein] |
-0.638 |
0.108 |
| 280 |
255794_at |
AT2G33480
|
ANAC041, NAC domain containing protein 41, NAC041, NAC domain containing protein 41 |
-0.636 |
-0.067 |
| 281 |
266940_at |
AT2G18970
|
unknown |
-0.636 |
0.050 |
| 282 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.635 |
0.241 |
| 283 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.626 |
0.538 |
| 284 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.626 |
-0.002 |
| 285 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.626 |
0.002 |
| 286 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.625 |
0.279 |
| 287 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
-0.624 |
0.389 |
| 288 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.622 |
-0.611 |
| 289 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
-0.619 |
0.249 |
| 290 |
263745_at |
AT2G21450
|
CHR34, chromatin remodeling 34 |
-0.616 |
-0.008 |
| 291 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
-0.614 |
0.828 |
| 292 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.613 |
0.473 |
| 293 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.613 |
-0.149 |
| 294 |
262531_at |
AT1G17230
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.610 |
0.006 |
| 295 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.603 |
0.005 |
| 296 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
-0.600 |
0.164 |
| 297 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
-0.599 |
0.449 |
| 298 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.596 |
0.654 |
| 299 |
249435_at |
AT5G39970
|
[catalytics] |
-0.594 |
-0.029 |
| 300 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-0.591 |
0.593 |
| 301 |
248260_at |
AT5G53240
|
unknown |
-0.588 |
-0.065 |
| 302 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.588 |
0.535 |
| 303 |
256524_at |
AT1G66200
|
ATGSR2, glutamine synthase clone F11, GLN1;2, glutamine synthetase 1;2, GSR2, glutamine synthase clone F11 |
-0.585 |
-0.107 |
| 304 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.584 |
0.612 |
| 305 |
249187_at |
AT5G43060
|
RD21B, esponsive to dehydration 21B |
-0.582 |
0.092 |
| 306 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.580 |
1.140 |
| 307 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
-0.578 |
-0.036 |
| 308 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.574 |
0.020 |
| 309 |
258354_at |
AT3G14320
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.571 |
-0.041 |
| 310 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.571 |
-0.466 |
| 311 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.568 |
-0.333 |
| 312 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
-0.567 |
0.605 |
| 313 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.563 |
-0.114 |
| 314 |
259909_at |
AT1G60870
|
MEE9, maternal effect embryo arrest 9 |
-0.561 |
-0.108 |
| 315 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.557 |
0.444 |
| 316 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.557 |
0.162 |
| 317 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.556 |
-0.707 |
| 318 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
-0.554 |
0.260 |
| 319 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.553 |
-0.772 |
| 320 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.551 |
0.014 |
| 321 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.543 |
0.405 |
| 322 |
251544_at |
AT3G58790
|
GAUT15, galacturonosyltransferase 15 |
-0.543 |
0.066 |
| 323 |
254786_at |
AT4G12890
|
[Gamma interferon responsive lysosomal thiol (GILT) reductase family protein] |
-0.541 |
0.006 |
| 324 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.540 |
-0.534 |
| 325 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
-0.540 |
0.915 |
| 326 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.539 |
0.041 |
| 327 |
245226_at |
AT3G29970
|
[B12D protein] |
-0.537 |
0.029 |
| 328 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.536 |
0.551 |
| 329 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
-0.530 |
-0.300 |
| 330 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
-0.530 |
0.540 |
| 331 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.530 |
0.137 |
| 332 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.527 |
-0.140 |
| 333 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
-0.524 |
-0.068 |
| 334 |
259015_at |
AT3G07350
|
unknown |
-0.523 |
0.249 |
| 335 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
-0.522 |
0.158 |
| 336 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.521 |
-0.184 |
| 337 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.520 |
-0.061 |
| 338 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.519 |
0.174 |
| 339 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.518 |
0.385 |
| 340 |
252751_at |
AT3G43430
|
[RING/U-box superfamily protein] |
-0.518 |
-0.012 |
| 341 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.517 |
-0.151 |
| 342 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.516 |
0.013 |
| 343 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.516 |
-0.232 |
| 344 |
246001_at |
AT5G20790
|
unknown |
-0.512 |
-0.646 |
| 345 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.511 |
-0.085 |
| 346 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.510 |
0.511 |
| 347 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
-0.510 |
0.031 |
| 348 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.509 |
-0.142 |
| 349 |
263258_at |
AT1G10540
|
ATNAT8, NAT8, nucleobase-ascorbate transporter 8 |
-0.508 |
0.015 |
| 350 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
-0.508 |
0.002 |
| 351 |
253965_at |
AT4G26490
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.503 |
-0.158 |
| 352 |
262096_at |
AT1G56010
|
anac021, Arabidopsis NAC domain containing protein 21, ANAC022, Arabidopsis NAC domain containing protein 22, NAC1, NAC domain containing protein 1 |
-0.502 |
0.056 |
| 353 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.501 |
-0.103 |
| 354 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.497 |
0.861 |
| 355 |
247884_at |
AT5G57800
|
CER3, ECERIFERUM 3, FLP1, FACELESS POLLEN 1, WAX2, YRE |
-0.496 |
0.042 |
| 356 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-0.491 |
-0.109 |
| 357 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-0.487 |
0.127 |
| 358 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.484 |
0.820 |
| 359 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.481 |
0.517 |