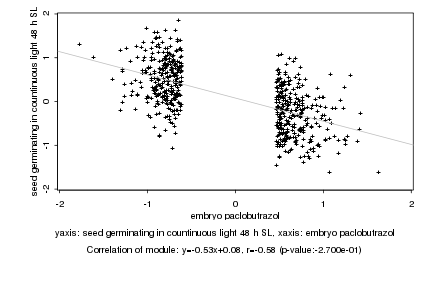
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
embryo paclobutrazol |
Log2 signal ratio
seed germinating in countinuous light 48 h SL |
| 1 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.619 |
-1.608 |
| 2 |
253895_at |
AT4G27160
|
AT2S3, SESA3, seed storage albumin 3 |
1.418 |
-0.272 |
| 3 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.407 |
-0.627 |
| 4 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.385 |
-0.908 |
| 5 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
1.302 |
0.610 |
| 6 |
257853_at |
AT3G12960
|
SMP1, seed maturation protein 1 |
1.270 |
-0.782 |
| 7 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
1.244 |
-0.889 |
| 8 |
266392_at |
AT2G41280
|
ATM10, M10 |
1.242 |
-0.966 |
| 9 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
1.237 |
0.337 |
| 10 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
1.234 |
-0.861 |
| 11 |
265095_at |
AT1G03880
|
CRB, CRUCIFERIN B, CRU2, cruciferin 2 |
1.222 |
-0.142 |
| 12 |
258327_at |
AT3G22640
|
PAP85 |
1.186 |
0.042 |
| 13 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
1.166 |
-0.890 |
| 14 |
253902_at |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
1.166 |
-1.165 |
| 15 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
1.132 |
-0.144 |
| 16 |
251731_at |
AT3G56350
|
[Iron/manganese superoxide dismutase family protein] |
1.126 |
-0.777 |
| 17 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
1.099 |
-0.137 |
| 18 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
1.083 |
-0.499 |
| 19 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
1.078 |
0.638 |
| 20 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
1.069 |
-0.823 |
| 21 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.066 |
-0.407 |
| 22 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.062 |
-1.613 |
| 23 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
1.045 |
-0.637 |
| 24 |
264580_at |
AT1G05340
|
unknown |
1.019 |
-0.877 |
| 25 |
263175_at |
AT1G05510
|
OBAP1a, Oil Body-Associated Protein 1a |
1.018 |
-0.862 |
| 26 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
1.017 |
-0.418 |
| 27 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
1.017 |
-0.129 |
| 28 |
267321_at |
AT2G19320
|
unknown |
1.008 |
-0.316 |
| 29 |
245070_at |
AT2G23240
|
AtMT4b, Arabidopsis thaliana metallothionein 4b |
0.991 |
0.101 |
| 30 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
0.986 |
0.109 |
| 31 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.985 |
-0.315 |
| 32 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.978 |
-0.370 |
| 33 |
265621_at |
AT2G27300
|
ANAC040, Arabidopsis NAC domain containing protein 40, NTL8, NTM1-like 8 |
0.962 |
-0.104 |
| 34 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
0.956 |
-0.567 |
| 35 |
260458_at |
AT1G68250
|
unknown |
0.953 |
-1.015 |
| 36 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.946 |
0.114 |
| 37 |
259314_at |
AT3G05260
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.944 |
-1.239 |
| 38 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.937 |
-0.960 |
| 39 |
253494_at |
AT4G31830
|
unknown |
0.931 |
-0.215 |
| 40 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.928 |
-0.995 |
| 41 |
251580_at |
AT3G58450
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.927 |
0.306 |
| 42 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.918 |
-0.072 |
| 43 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
0.918 |
-0.489 |
| 44 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
0.902 |
0.058 |
| 45 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.892 |
-0.385 |
| 46 |
248915_at |
AT5G45690
|
unknown |
0.884 |
-1.134 |
| 47 |
255493_at |
AT4G02690
|
AtLFG3, LFG3, LIFEGUARD 3 |
0.878 |
-1.071 |
| 48 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
0.873 |
-0.089 |
| 49 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.867 |
-0.781 |
| 50 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.864 |
-0.909 |
| 51 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.862 |
-0.566 |
| 52 |
260459_at |
AT1G68240
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.858 |
-0.588 |
| 53 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
0.856 |
-0.143 |
| 54 |
247421_at |
AT5G62800
|
[Protein with RING/U-box and TRAF-like domains] |
0.851 |
-0.864 |
| 55 |
260761_at |
AT1G49150
|
unknown |
0.849 |
-1.090 |
| 56 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.836 |
-0.925 |
| 57 |
262527_at |
AT1G17010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.834 |
-1.286 |
| 58 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.829 |
0.118 |
| 59 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.824 |
-1.234 |
| 60 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.819 |
-0.114 |
| 61 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.815 |
-0.375 |
| 62 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
0.814 |
-0.499 |
| 63 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
0.814 |
-1.161 |
| 64 |
259782_at |
AT1G29680
|
unknown |
0.806 |
0.143 |
| 65 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
0.797 |
-0.536 |
| 66 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
0.794 |
-0.427 |
| 67 |
255980_at |
AT1G33970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.792 |
0.511 |
| 68 |
265094_at |
AT1G03890
|
[RmlC-like cupins superfamily protein] |
0.785 |
-0.142 |
| 69 |
253322_at |
AT4G33980
|
unknown |
0.778 |
-1.242 |
| 70 |
247714_at |
AT5G59340
|
WOX2, WUSCHEL related homeobox 2 |
0.773 |
-1.389 |
| 71 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.771 |
-0.653 |
| 72 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.769 |
-0.856 |
| 73 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
0.766 |
-0.582 |
| 74 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.765 |
-0.707 |
| 75 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.760 |
-1.011 |
| 76 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.758 |
0.093 |
| 77 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.757 |
0.027 |
| 78 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.755 |
-0.088 |
| 79 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.755 |
-0.165 |
| 80 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
0.754 |
-0.043 |
| 81 |
247172_at |
AT5G65550
|
[UDP-Glycosyltransferase superfamily protein] |
0.753 |
0.380 |
| 82 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
0.752 |
-0.196 |
| 83 |
248096_at |
AT5G55240
|
ATPXG2, ARABIDOPSIS THALIANA PEROXYGENASE 2 |
0.746 |
-0.628 |
| 84 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.745 |
-0.257 |
| 85 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.744 |
-0.241 |
| 86 |
245233_at |
AT4G25580
|
[CAP160 protein] |
0.743 |
-0.259 |
| 87 |
250468_at |
AT5G10120
|
[Ethylene insensitive 3 family protein] |
0.742 |
-0.632 |
| 88 |
247514_at |
AT5G61640
|
ATMSRA1, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE A1, PMSR1, peptidemethionine sulfoxide reductase 1 |
0.738 |
-0.162 |
| 89 |
249937_at |
AT5G22470
|
[NAD+ ADP-ribosyltransferases] |
0.735 |
-0.327 |
| 90 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
0.734 |
-0.610 |
| 91 |
262185_at |
AT1G77950
|
AGL67, AGAMOUS-like 67 |
0.733 |
-0.667 |
| 92 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
0.733 |
-0.361 |
| 93 |
249438_at |
AT5G40010
|
AATP1, AAA-ATPase 1, ASD, ATPase-in-Seed-Development |
0.731 |
-1.278 |
| 94 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
0.730 |
0.785 |
| 95 |
248076_at |
AT5G55750
|
[hydroxyproline-rich glycoprotein family protein] |
0.730 |
-0.277 |
| 96 |
267579_at |
AT2G42000
|
AtMT4a, Arabidopsis thaliana metallothionein 4a |
0.728 |
-0.090 |
| 97 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.728 |
0.328 |
| 98 |
255840_at |
AT2G33520
|
unknown |
0.725 |
-0.643 |
| 99 |
265084_at |
AT1G03790
|
AtTZF4, SOM, SOMNUS, TZF4, Tandem CCCH Zinc Finger protein 4 |
0.722 |
-0.625 |
| 100 |
265891_at |
AT2G15010
|
[Plant thionin] |
0.722 |
0.505 |
| 101 |
265276_at |
AT2G28400
|
unknown |
0.719 |
0.012 |
| 102 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.715 |
0.422 |
| 103 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.708 |
0.049 |
| 104 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.707 |
0.637 |
| 105 |
255763_at |
AT1G16730
|
unknown |
0.707 |
-0.768 |
| 106 |
265099_at |
AT1G03990
|
[Long-chain fatty alcohol dehydrogenase family protein] |
0.705 |
-0.061 |
| 107 |
246018_at |
AT5G10695
|
unknown |
0.702 |
-0.303 |
| 108 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.695 |
0.324 |
| 109 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.695 |
-0.120 |
| 110 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.694 |
-0.887 |
| 111 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.690 |
0.075 |
| 112 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.686 |
-0.205 |
| 113 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
0.684 |
-0.972 |
| 114 |
247953_at |
AT5G57260
|
CYP71B10, cytochrome P450, family 71, subfamily B, polypeptide 10 |
0.683 |
-0.910 |
| 115 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.679 |
-0.935 |
| 116 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.678 |
0.986 |
| 117 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.677 |
-0.195 |
| 118 |
260005_at |
AT1G67920
|
unknown |
0.676 |
0.179 |
| 119 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.675 |
-0.004 |
| 120 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.673 |
-0.810 |
| 121 |
255471_at |
AT4G03050
|
AOP3 |
0.673 |
0.556 |
| 122 |
250426_at |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
0.671 |
-0.509 |
| 123 |
247196_at |
AT5G65510
|
AIL7, AINTEGUMENTA-like 7, PLT7, PLETHORA 7 |
0.670 |
-0.714 |
| 124 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.669 |
-0.577 |
| 125 |
255049_at |
AT4G09610
|
GASA2, GAST1 protein homolog 2 |
0.663 |
-0.720 |
| 126 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.661 |
-0.068 |
| 127 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.661 |
-0.336 |
| 128 |
249688_at |
AT5G36160
|
[Tyrosine transaminase family protein] |
0.659 |
0.923 |
| 129 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
0.656 |
0.288 |
| 130 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.653 |
-0.799 |
| 131 |
262378_at |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
0.651 |
-0.309 |
| 132 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.651 |
0.289 |
| 133 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.640 |
-0.311 |
| 134 |
247269_at |
AT5G64210
|
AOX2, alternative oxidase 2 |
0.639 |
-0.557 |
| 135 |
261496_at |
AT1G28360
|
ATERF12, ERF DOMAIN PROTEIN 12, ERF12, ERF domain protein 12 |
0.638 |
-0.646 |
| 136 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
0.637 |
-1.078 |
| 137 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
0.636 |
-0.198 |
| 138 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.629 |
-0.370 |
| 139 |
253870_at |
AT4G27530
|
unknown |
0.624 |
-0.803 |
| 140 |
261586_at |
AT1G01640
|
[BTB/POZ domain-containing protein] |
0.623 |
-0.015 |
| 141 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
0.622 |
-0.232 |
| 142 |
253796_at |
AT4G28460
|
unknown |
0.619 |
0.497 |
| 143 |
267313_at |
AT2G34740
|
[Protein phosphatase 2C family protein] |
0.617 |
-1.097 |
| 144 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.616 |
0.068 |
| 145 |
255461_at |
AT4G02780
|
ABC33, ATCPS1, ARABIDOPSIS THALIANA ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, CPS, CPP synthase, CPS1, ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, GA1, GA REQUIRING 1 |
0.615 |
0.190 |
| 146 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
0.614 |
-0.840 |
| 147 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
0.614 |
-0.509 |
| 148 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
0.613 |
0.110 |
| 149 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
0.607 |
1.001 |
| 150 |
250010_at |
AT5G18450
|
[Integrase-type DNA-binding superfamily protein] |
0.605 |
0.407 |
| 151 |
257074_at |
AT3G19660
|
unknown |
0.605 |
-0.364 |
| 152 |
263847_at |
AT2G36970
|
[UDP-Glycosyltransferase superfamily protein] |
0.603 |
0.465 |
| 153 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
0.602 |
-0.087 |
| 154 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.597 |
-1.150 |
| 155 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.596 |
0.392 |
| 156 |
255900_at |
AT1G17830
|
unknown |
0.595 |
0.550 |
| 157 |
247226_at |
AT5G65100
|
[Ethylene insensitive 3 family protein] |
0.595 |
-0.439 |
| 158 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.595 |
-0.086 |
| 159 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
0.594 |
-0.060 |
| 160 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
0.592 |
-0.689 |
| 161 |
257122_at |
AT3G20250
|
APUM5, pumilio 5, PUM5, pumilio 5 |
0.592 |
-0.147 |
| 162 |
252914_at |
AT4G39130
|
[Dehydrin family protein] |
0.589 |
-0.679 |
| 163 |
261866_at |
AT1G50420
|
SCL-3, SCARECROW-LIKE 3, SCL3, scarecrow-like 3 |
0.587 |
0.009 |
| 164 |
249068_at |
AT5G43980
|
PDLP1, plasmodesmata-located protein 1, PDLP1A, PLASMODESMATA-LOCATED PROTEIN 1A |
0.586 |
-0.637 |
| 165 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.586 |
-0.806 |
| 166 |
265714_at |
AT2G03520
|
ATUPS4, ureide permease 4, UPS4, ureide permease 4 |
0.586 |
-0.077 |
| 167 |
264726_at |
AT1G22985
|
CRF7, cytokinin response factor 7 |
0.585 |
-0.496 |
| 168 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.579 |
-0.426 |
| 169 |
259578_at |
AT1G27990
|
unknown |
0.577 |
-0.755 |
| 170 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.577 |
-0.202 |
| 171 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.577 |
-0.792 |
| 172 |
253305_at |
AT4G33666
|
unknown |
0.577 |
-0.849 |
| 173 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.576 |
-0.189 |
| 174 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
0.575 |
-0.173 |
| 175 |
255452_at |
AT4G02880
|
unknown |
0.575 |
0.852 |
| 176 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
0.575 |
0.275 |
| 177 |
261936_at |
AT1G22600
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.574 |
-0.504 |
| 178 |
266540_at |
AT2G35310
|
[Transcriptional factor B3 family protein] |
0.572 |
-0.838 |
| 179 |
253984_at |
AT4G26590
|
ATOPT5, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 5, OPT5, oligopeptide transporter 5 |
0.570 |
-0.187 |
| 180 |
263931_at |
AT2G36220
|
unknown |
0.566 |
0.632 |
| 181 |
260917_at |
AT1G02700
|
unknown |
0.566 |
0.258 |
| 182 |
251245_at |
AT3G62090
|
PIF6, PHYTOCHROME-INTERACTING FACTOR 6, PIL2, phytochrome interacting factor 3-like 2 |
0.565 |
0.027 |
| 183 |
256114_at |
AT1G16850
|
unknown |
0.564 |
0.158 |
| 184 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.563 |
-0.039 |
| 185 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.563 |
-0.730 |
| 186 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
0.561 |
-0.900 |
| 187 |
260668_at |
AT1G19530
|
unknown |
0.560 |
0.158 |
| 188 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.559 |
0.679 |
| 189 |
254391_at |
AT4G21590
|
ENDO3, endonuclease 3 |
0.559 |
-1.138 |
| 190 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.557 |
-0.834 |
| 191 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.557 |
-0.977 |
| 192 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.554 |
-0.525 |
| 193 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.553 |
-0.338 |
| 194 |
250438_at |
AT5G10580
|
unknown |
0.552 |
0.452 |
| 195 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.550 |
0.476 |
| 196 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.547 |
0.394 |
| 197 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.546 |
-0.256 |
| 198 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
0.546 |
0.106 |
| 199 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.545 |
-0.698 |
| 200 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
0.544 |
0.071 |
| 201 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.542 |
-0.092 |
| 202 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.541 |
0.372 |
| 203 |
255527_at |
AT4G02360
|
unknown |
0.540 |
-0.634 |
| 204 |
256057_at |
AT1G07180
|
ATNDI1, ARABIDOPSIS THALIANA INTERNAL NON-PHOSPHORYLATING NAD ( P ) H DEHYDROGENASE, NDA1, alternative NAD(P)H dehydrogenase 1 |
0.539 |
0.034 |
| 205 |
246332_at |
AT3G44830
|
[Lecithin:cholesterol acyltransferase family protein] |
0.536 |
0.083 |
| 206 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.536 |
-0.888 |
| 207 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
0.535 |
0.236 |
| 208 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.532 |
-0.250 |
| 209 |
254630_at |
AT4G18360
|
GOX3, glycolate oxidase 3 |
0.531 |
0.433 |
| 210 |
248969_at |
AT5G45310
|
unknown |
0.530 |
-0.892 |
| 211 |
263877_at |
AT2G21780
|
unknown |
0.528 |
-1.026 |
| 212 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
0.527 |
0.404 |
| 213 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
0.527 |
-0.748 |
| 214 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
0.525 |
-0.163 |
| 215 |
247177_at |
AT5G65300
|
unknown |
0.524 |
0.115 |
| 216 |
265266_at |
AT2G42890
|
AML2, MEI2-like 2, ML2, MEI2-like 2 |
0.524 |
-0.064 |
| 217 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.523 |
-0.638 |
| 218 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.523 |
-0.178 |
| 219 |
254691_at |
AT4G17840
|
unknown |
0.523 |
-0.385 |
| 220 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.521 |
0.439 |
| 221 |
257869_at |
AT3G25160
|
[ER lumen protein retaining receptor family protein] |
0.521 |
0.315 |
| 222 |
256898_at |
AT3G24650
|
ABI3, ABA INSENSITIVE 3, ABI3, ABSCISIC ACID INSENSITIVE 3, SIS10, SUGAR INSENSITIVE 10 |
0.521 |
-0.716 |
| 223 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.519 |
-0.267 |
| 224 |
249255_at |
AT5G41610
|
ATCHX18, ARABIDOPSIS THALIANA CATION/H+ EXCHANGER 18, CHX18, cation/H+ exchanger 18 |
0.518 |
-0.499 |
| 225 |
254724_at |
AT4G13630
|
unknown |
0.517 |
1.080 |
| 226 |
249746_at |
AT5G24590
|
ANAC091, Arabidopsis NAC domain containing protein 91, TIP, TCV-interacting protein |
0.517 |
0.404 |
| 227 |
262295_at |
AT1G27650
|
ATU2AF35A |
0.514 |
-0.146 |
| 228 |
260878_at |
AT1G21450
|
SCL1, SCARECROW-like 1 |
0.514 |
0.350 |
| 229 |
257466_at |
AT1G62840
|
unknown |
0.514 |
-0.637 |
| 230 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
0.513 |
-0.602 |
| 231 |
258257_at |
AT3G26770
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.513 |
0.188 |
| 232 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
0.513 |
-0.604 |
| 233 |
262478_at |
AT1G11170
|
unknown |
0.511 |
-0.290 |
| 234 |
251202_at |
AT3G63040
|
unknown |
0.511 |
-0.980 |
| 235 |
265828_at |
AT2G14520
|
unknown |
0.511 |
-0.173 |
| 236 |
252173_at |
AT3G50650
|
[GRAS family transcription factor] |
0.509 |
-0.578 |
| 237 |
264024_at |
AT2G21180
|
unknown |
0.509 |
-0.174 |
| 238 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.508 |
0.221 |
| 239 |
263881_at |
AT2G21820
|
unknown |
0.508 |
-0.867 |
| 240 |
254200_at |
AT4G24110
|
unknown |
0.507 |
-0.538 |
| 241 |
262607_at |
AT1G13990
|
unknown |
0.506 |
-0.409 |
| 242 |
264352_at |
AT1G03270
|
unknown |
0.503 |
0.099 |
| 243 |
256777_at |
AT3G13780
|
[SMAD/FHA domain-containing protein ] |
0.503 |
0.531 |
| 244 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
0.500 |
0.150 |
| 245 |
250501_at |
AT5G09640
|
SCPL19, serine carboxypeptidase-like 19, SNG2, SINAPOYLGLUCOSE ACCUMULATOR 2 |
0.500 |
-1.244 |
| 246 |
254917_at |
AT4G11350
|
unknown |
0.499 |
0.369 |
| 247 |
247128_at |
AT5G66110
|
HIPP27, heavy metal associated isoprenylated plant protein 27 |
0.499 |
-0.494 |
| 248 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.497 |
-0.749 |
| 249 |
257369_at |
AT2G35550
|
ATBPC7, BASIC PENTACYSTEINE 7, BBR, BPC7, basic pentacysteine 7 |
0.497 |
-1.016 |
| 250 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.496 |
-0.479 |
| 251 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.495 |
0.673 |
| 252 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
0.494 |
-0.005 |
| 253 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.493 |
-0.403 |
| 254 |
248495_at |
AT5G50780
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.493 |
-0.337 |
| 255 |
258263_at |
AT3G15780
|
unknown |
0.493 |
-1.275 |
| 256 |
262322_at |
AT1G27590
|
unknown |
0.492 |
-0.687 |
| 257 |
255283_at |
AT4G04620
|
ATG8B, autophagy 8b |
0.491 |
-0.479 |
| 258 |
262977_at |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
0.491 |
-0.360 |
| 259 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.490 |
0.504 |
| 260 |
258939_at |
AT3G10020
|
unknown |
0.489 |
0.139 |
| 261 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.488 |
-0.611 |
| 262 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
0.487 |
0.627 |
| 263 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
0.485 |
1.067 |
| 264 |
250118_at |
AT5G16460
|
[Putative adipose-regulatory protein (Seipin)] |
0.485 |
-0.916 |
| 265 |
257859_at |
AT3G12955
|
SAUR74, SMALL AUXIN UPREGULATED RNA 74 |
0.483 |
-0.922 |
| 266 |
253776_at |
AT4G28390
|
AAC3, ADP/ATP carrier 3, ATAAC3 |
0.483 |
0.116 |
| 267 |
251187_at |
AT3G62770
|
AtATG18a, autophagy 18a, ATG18a, autophagy 18a, PEUP2, PEROXISOME UNUSUAL POSITIONING 2 |
0.482 |
1.064 |
| 268 |
260635_at |
AT1G62422
|
unknown |
0.482 |
-0.091 |
| 269 |
255602_at |
AT4G01026
|
PYL7, PYR1-like 7, RCAR2, regulatory components of ABA receptor 2 |
0.481 |
0.734 |
| 270 |
259178_at |
AT3G01650
|
RGLG1, RING domain ligase1 |
0.481 |
-0.121 |
| 271 |
256938_at |
AT3G22500
|
ATECP31, LATE EMBRYOGENESIS ABUNDANT PROTEIN ECP31 |
0.480 |
0.062 |
| 272 |
247883_at |
AT5G57790
|
unknown |
0.480 |
-0.776 |
| 273 |
267036_at |
AT2G38465
|
unknown |
0.479 |
-0.587 |
| 274 |
257224_at |
AT3G27870
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.479 |
0.520 |
| 275 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.478 |
0.340 |
| 276 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.477 |
-0.782 |
| 277 |
267286_at |
AT2G23640
|
RTNLB13, Reticulan like protein B13 |
0.477 |
-0.103 |
| 278 |
257309_at |
AT3G28150
|
AXY4L, ALTERED XYLOGLUCAN 4-LIKE, TBL22, TRICHOME BIREFRINGENCE-LIKE 22 |
0.477 |
-0.249 |
| 279 |
265086_at |
AT1G03770
|
ATRING1B, ARABIDOPSIS THALIANA RING 1B, RING1B, RING 1B |
0.473 |
-0.965 |
| 280 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.472 |
0.507 |
| 281 |
250555_at |
AT5G07950
|
unknown |
0.472 |
-0.128 |
| 282 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.472 |
0.250 |
| 283 |
245650_at |
AT1G24735
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.472 |
0.728 |
| 284 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
0.471 |
-1.015 |
| 285 |
261973_at |
AT1G64610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.471 |
0.318 |
| 286 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.471 |
-0.295 |
| 287 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.470 |
-0.477 |
| 288 |
251882_at |
AT3G54140
|
AtNPF8.1, ATPTR1, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 1, NPF8.1, NRT1/ PTR family 8.1, PTR1, peptide transporter 1 |
0.470 |
0.471 |
| 289 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.469 |
-0.531 |
| 290 |
246951_at |
AT5G04880
|
[expressed protein] |
0.468 |
-1.007 |
| 291 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
0.468 |
0.461 |
| 292 |
250028_at |
AT5G18130
|
unknown |
0.466 |
-0.172 |
| 293 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.466 |
0.324 |
| 294 |
257451_at |
AT1G05690
|
BT3, BTB and TAZ domain protein 3 |
0.466 |
0.453 |
| 295 |
262255_at |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
0.465 |
-0.896 |
| 296 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.465 |
-0.006 |
| 297 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.465 |
-1.441 |
| 298 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.464 |
-0.540 |
| 299 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.464 |
0.112 |
| 300 |
246743_at |
AT5G27750
|
[F-box/FBD-like domains containing protein] |
0.461 |
0.052 |
| 301 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.775 |
1.322 |
| 302 |
254234_at |
AT4G23680
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.623 |
1.012 |
| 303 |
257313_at |
AT3G26520
|
GAMMA-TIP2, SITIP, SALT-STRESS INDUCIBLE TONOPLAST INTRINSIC PROTEIN, TIP1;2, TIP2, tonoplast intrinsic protein 2 |
-1.407 |
0.518 |
| 304 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-1.312 |
1.182 |
| 305 |
247878_at |
AT5G57760
|
unknown |
-1.310 |
-0.187 |
| 306 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.294 |
-0.009 |
| 307 |
262236_at |
AT1G48330
|
unknown |
-1.293 |
0.691 |
| 308 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
-1.288 |
0.742 |
| 309 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.271 |
0.394 |
| 310 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-1.264 |
0.132 |
| 311 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.249 |
1.218 |
| 312 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-1.200 |
0.933 |
| 313 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-1.186 |
0.427 |
| 314 |
253814_at |
AT4G28290
|
unknown |
-1.183 |
0.150 |
| 315 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-1.178 |
0.233 |
| 316 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
-1.143 |
0.485 |
| 317 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-1.141 |
-0.168 |
| 318 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-1.133 |
1.264 |
| 319 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-1.119 |
0.160 |
| 320 |
259788_at |
AT1G29670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.118 |
0.179 |
| 321 |
265118_at |
AT1G62660
|
[Glycosyl hydrolases family 32 protein] |
-1.117 |
1.006 |
| 322 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.085 |
0.439 |
| 323 |
254328_at |
AT4G22570
|
APT3, adenine phosphoribosyl transferase 3 |
-1.076 |
0.332 |
| 324 |
266123_at |
AT2G45180
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.072 |
1.252 |
| 325 |
247768_at |
AT5G58900
|
[Homeodomain-like transcriptional regulator] |
-1.065 |
0.722 |
| 326 |
258751_at |
AT3G05890
|
RCI2B, RARE-COLD-INDUCIBLE 2B |
-1.059 |
0.654 |
| 327 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-1.049 |
0.944 |
| 328 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-1.048 |
1.360 |
| 329 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-1.035 |
1.021 |
| 330 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-1.033 |
0.678 |
| 331 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-1.031 |
0.724 |
| 332 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-1.029 |
1.168 |
| 333 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-1.019 |
1.676 |
| 334 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-1.015 |
0.672 |
| 335 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-1.012 |
0.088 |
| 336 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-1.005 |
0.127 |
| 337 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-1.003 |
-0.007 |
| 338 |
248681_at |
AT5G48900
|
[Pectin lyase-like superfamily protein] |
-0.999 |
0.759 |
| 339 |
250891_at |
AT5G04530
|
KCS19, 3-ketoacyl-CoA synthase 19 |
-0.997 |
0.114 |
| 340 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.993 |
0.795 |
| 341 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.991 |
-0.313 |
| 342 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.974 |
0.629 |
| 343 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.969 |
-0.343 |
| 344 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.967 |
0.796 |
| 345 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.965 |
0.544 |
| 346 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.964 |
1.014 |
| 347 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.962 |
0.958 |
| 348 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.959 |
0.502 |
| 349 |
246004_at |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
-0.959 |
1.227 |
| 350 |
256926_at |
AT3G22540
|
unknown |
-0.957 |
0.085 |
| 351 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
-0.950 |
1.112 |
| 352 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.949 |
1.358 |
| 353 |
247477_at |
AT5G62340
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.949 |
0.332 |
| 354 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.935 |
0.260 |
| 355 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.932 |
0.259 |
| 356 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
-0.930 |
0.203 |
| 357 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.929 |
1.585 |
| 358 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
-0.928 |
0.289 |
| 359 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.927 |
0.697 |
| 360 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.925 |
0.318 |
| 361 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.924 |
0.657 |
| 362 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.924 |
0.971 |
| 363 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.923 |
0.540 |
| 364 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.923 |
-0.588 |
| 365 |
247645_at |
AT5G60530
|
[late embryogenesis abundant protein-related / LEA protein-related] |
-0.920 |
0.335 |
| 366 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.920 |
0.197 |
| 367 |
259996_at |
AT1G67910
|
unknown |
-0.918 |
0.445 |
| 368 |
265296_at |
AT2G14060
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.916 |
-0.106 |
| 369 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.915 |
1.461 |
| 370 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.909 |
-0.278 |
| 371 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.908 |
0.259 |
| 372 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.907 |
1.265 |
| 373 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.906 |
0.283 |
| 374 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-0.904 |
0.117 |
| 375 |
250778_at |
AT5G05500
|
MOP10 |
-0.902 |
1.483 |
| 376 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.900 |
0.358 |
| 377 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.899 |
1.454 |
| 378 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.896 |
0.541 |
| 379 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.896 |
1.584 |
| 380 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.895 |
0.020 |
| 381 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.893 |
0.159 |
| 382 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.888 |
-0.033 |
| 383 |
261256_at |
AT1G05760
|
RTM1, restricted tev movement 1 |
-0.884 |
0.484 |
| 384 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.880 |
0.162 |
| 385 |
263737_at |
AT1G60010
|
unknown |
-0.880 |
0.154 |
| 386 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.880 |
0.516 |
| 387 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.879 |
-0.061 |
| 388 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.878 |
0.410 |
| 389 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.878 |
1.132 |
| 390 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.876 |
1.304 |
| 391 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
-0.873 |
-0.754 |
| 392 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.872 |
1.250 |
| 393 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.870 |
0.762 |
| 394 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.870 |
0.684 |
| 395 |
248341_at |
AT5G52220
|
unknown |
-0.870 |
0.379 |
| 396 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.866 |
1.475 |
| 397 |
254983_at |
AT4G10560
|
MEE53, maternal effect embryo arrest 53 |
-0.866 |
-0.273 |
| 398 |
258468_at |
AT3G06070
|
unknown |
-0.865 |
-0.792 |
| 399 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.865 |
0.919 |
| 400 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
-0.863 |
0.384 |
| 401 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.859 |
0.748 |
| 402 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
-0.856 |
0.184 |
| 403 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.852 |
0.596 |
| 404 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
-0.842 |
0.316 |
| 405 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.841 |
0.338 |
| 406 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.840 |
0.099 |
| 407 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.840 |
1.365 |
| 408 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.839 |
0.127 |
| 409 |
245172_at |
AT2G47540
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.838 |
0.775 |
| 410 |
267636_at |
AT2G42110
|
unknown |
-0.838 |
-0.003 |
| 411 |
255720_at |
AT1G32060
|
PRK, phosphoribulokinase |
-0.835 |
0.943 |
| 412 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.835 |
0.425 |
| 413 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.834 |
0.658 |
| 414 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.832 |
0.101 |
| 415 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
-0.824 |
0.828 |
| 416 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.823 |
0.720 |
| 417 |
250059_at |
AT5G17820
|
[Peroxidase superfamily protein] |
-0.822 |
0.544 |
| 418 |
267392_at |
AT2G44490
|
BGLU26, BETA GLUCOSIDASE 26, PEN2, PENETRATION 2 |
-0.820 |
1.139 |
| 419 |
248186_at |
AT5G53880
|
unknown |
-0.816 |
0.459 |
| 420 |
249847_at |
AT5G23210
|
SCPL34, serine carboxypeptidase-like 34 |
-0.815 |
0.235 |
| 421 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
-0.815 |
0.053 |
| 422 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.812 |
0.990 |
| 423 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.812 |
0.224 |
| 424 |
261691_at |
AT1G50060
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.811 |
0.118 |
| 425 |
257151_at |
AT3G27200
|
[Cupredoxin superfamily protein] |
-0.809 |
0.144 |
| 426 |
265029_at |
AT1G24625
|
ZFP7, zinc finger protein 7 |
-0.809 |
0.228 |
| 427 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.807 |
0.885 |
| 428 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.805 |
0.966 |
| 429 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.804 |
0.797 |
| 430 |
267154_at |
AT2G30870
|
ATGSTF10, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 10, ATGSTF4, ERD13, EARLY DEHYDRATION-INDUCED 13, GSTF10, glutathione S-transferase PHI 10 |
-0.804 |
1.631 |
| 431 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.803 |
1.213 |
| 432 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.803 |
-0.344 |
| 433 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.802 |
0.767 |
| 434 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
-0.801 |
1.037 |
| 435 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.800 |
0.874 |
| 436 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
-0.800 |
-0.643 |
| 437 |
264313_at |
AT1G70410
|
ATBCA4, BETA CARBONIC ANHYDRASE 4, BCA4, beta carbonic anhydrase 4, CA4, BETA CARBONIC ANHYDRASE 4 |
-0.799 |
0.026 |
| 438 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
-0.796 |
0.658 |
| 439 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.796 |
0.849 |
| 440 |
264687_at |
AT1G09850
|
XBCP3, xylem bark cysteine peptidase 3 |
-0.793 |
0.974 |
| 441 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
-0.790 |
0.184 |
| 442 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.790 |
0.078 |
| 443 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.790 |
0.801 |
| 444 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.789 |
0.260 |
| 445 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.788 |
-0.032 |
| 446 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-0.785 |
0.980 |
| 447 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.784 |
-0.339 |
| 448 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.781 |
0.682 |
| 449 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
-0.781 |
1.132 |
| 450 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.779 |
1.065 |
| 451 |
264998_at |
AT1G67330
|
unknown |
-0.777 |
0.850 |
| 452 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.777 |
0.579 |
| 453 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.777 |
-0.053 |
| 454 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
-0.777 |
0.124 |
| 455 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.776 |
1.225 |
| 456 |
249836_at |
AT5G23420
|
HMGB6, high-mobility group box 6 |
-0.774 |
0.551 |
| 457 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.773 |
0.026 |
| 458 |
245076_at |
AT2G23170
|
GH3.3 |
-0.773 |
0.392 |
| 459 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.771 |
0.489 |
| 460 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.769 |
0.469 |
| 461 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.768 |
0.259 |
| 462 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.768 |
0.411 |
| 463 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
-0.766 |
0.496 |
| 464 |
263137_at |
AT1G78660
|
ATGGH1, gamma-glutamyl hydrolase 1, GGH1, gamma-glutamyl hydrolase 1 |
-0.764 |
0.853 |
| 465 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.762 |
-0.155 |
| 466 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.760 |
1.115 |
| 467 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.757 |
0.573 |
| 468 |
252058_at |
AT3G52470
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.757 |
0.915 |
| 469 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.754 |
-0.331 |
| 470 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.751 |
-0.406 |
| 471 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.751 |
0.991 |
| 472 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.748 |
1.461 |
| 473 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.746 |
-0.180 |
| 474 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.746 |
1.091 |
| 475 |
267299_at |
AT2G30150
|
[UDP-Glycosyltransferase superfamily protein] |
-0.746 |
-0.015 |
| 476 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-0.745 |
1.269 |
| 477 |
264577_at |
AT1G05260
|
RCI3, RARE COLD INDUCIBLE GENE 3, RCI3A |
-0.744 |
-0.462 |
| 478 |
262796_at |
AT1G20850
|
XCP2, xylem cysteine peptidase 2 |
-0.741 |
0.854 |
| 479 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.740 |
0.580 |
| 480 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.738 |
0.249 |
| 481 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.738 |
0.731 |
| 482 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.736 |
-0.035 |
| 483 |
263711_at |
AT2G20630
|
PIA1, PP2C induced by AVRRPM1 |
-0.736 |
0.307 |
| 484 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
-0.736 |
0.739 |
| 485 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.735 |
0.744 |
| 486 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.734 |
0.178 |
| 487 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
-0.734 |
0.174 |
| 488 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.734 |
-0.058 |
| 489 |
266271_at |
AT2G29440
|
ATGSTU6, glutathione S-transferase tau 6, GST24, GLUTATHIONE S-TRANSFERASE 24, GSTU6, glutathione S-transferase tau 6 |
-0.733 |
-0.030 |
| 490 |
258091_at |
AT3G14560
|
unknown |
-0.733 |
0.349 |
| 491 |
245309_at |
AT4G15140
|
unknown |
-0.733 |
0.235 |
| 492 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.732 |
0.029 |
| 493 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.732 |
0.709 |
| 494 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.732 |
0.210 |
| 495 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.729 |
0.778 |
| 496 |
263597_at |
AT2G01870
|
unknown |
-0.727 |
1.046 |
| 497 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.727 |
0.217 |
| 498 |
263481_at |
AT2G04025
|
GLV7, GOLVEN 7, RGF3, root meristem growth factor 3 |
-0.726 |
-0.223 |
| 499 |
249919_at |
AT5G19250
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.725 |
0.675 |
| 500 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.724 |
0.763 |
| 501 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.719 |
-1.055 |
| 502 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.719 |
-0.482 |
| 503 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.717 |
0.924 |
| 504 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
-0.716 |
0.912 |
| 505 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.715 |
0.261 |
| 506 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.714 |
0.982 |
| 507 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
-0.714 |
0.643 |
| 508 |
266884_at |
AT2G44790
|
UCC2, uclacyanin 2 |
-0.712 |
0.935 |
| 509 |
247474_at |
AT5G62280
|
unknown |
-0.712 |
0.086 |
| 510 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.712 |
1.072 |
| 511 |
246993_at |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-0.709 |
-0.079 |
| 512 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
-0.708 |
0.462 |
| 513 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.707 |
0.956 |
| 514 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.706 |
0.606 |
| 515 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.705 |
0.656 |
| 516 |
254392_at |
AT4G21600
|
ENDO5, endonuclease 5 |
-0.703 |
-0.184 |
| 517 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.702 |
0.091 |
| 518 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.700 |
-0.564 |
| 519 |
254248_at |
AT4G23270
|
CRK19, cysteine-rich RLK (RECEPTOR-like protein kinase) 19 |
-0.700 |
0.844 |
| 520 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.700 |
-0.286 |
| 521 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
-0.698 |
0.577 |
| 522 |
258530_at |
AT3G06840
|
unknown |
-0.698 |
0.059 |
| 523 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.694 |
0.170 |
| 524 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.693 |
0.432 |
| 525 |
246093_at |
AT5G20550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.693 |
0.467 |
| 526 |
265400_at |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.692 |
0.759 |
| 527 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.691 |
1.632 |
| 528 |
249887_at |
AT5G22310
|
unknown |
-0.690 |
0.866 |
| 529 |
249970_at |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
-0.690 |
-0.371 |
| 530 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-0.689 |
0.620 |
| 531 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.689 |
0.613 |
| 532 |
261970_at |
AT1G65960
|
GAD2, glutamate decarboxylase 2 |
-0.689 |
0.473 |
| 533 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
-0.687 |
0.056 |
| 534 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
-0.687 |
0.875 |
| 535 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.686 |
0.380 |
| 536 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.684 |
0.154 |
| 537 |
250881_at |
AT5G04080
|
unknown |
-0.683 |
-0.726 |
| 538 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.683 |
0.368 |
| 539 |
248028_at |
AT5G55620
|
unknown |
-0.682 |
0.093 |
| 540 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.681 |
0.472 |
| 541 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.679 |
0.607 |
| 542 |
249167_at |
AT5G42860
|
unknown |
-0.677 |
1.175 |
| 543 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.677 |
0.505 |
| 544 |
266244_at |
AT2G27740
|
unknown |
-0.677 |
-0.187 |
| 545 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.676 |
1.151 |
| 546 |
261999_at |
AT1G33800
|
AtGXMT1, GXM1, glucuronoxylan methyltransferase 1, GXMT1, glucuronoxylan methyltransferase 1 |
-0.674 |
0.419 |
| 547 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.673 |
0.195 |
| 548 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.671 |
0.615 |
| 549 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
-0.671 |
0.132 |
| 550 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
-0.670 |
1.437 |
| 551 |
258367_at |
AT3G14370
|
WAG2 |
-0.670 |
0.155 |
| 552 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.670 |
0.672 |
| 553 |
245701_at |
AT5G04140
|
FD-GOGAT, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE, GLS1, FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1, GLU1, glutamate synthase 1, GLUS |
-0.669 |
0.334 |
| 554 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.669 |
0.186 |
| 555 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.669 |
0.915 |
| 556 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.667 |
0.539 |
| 557 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
-0.667 |
-0.190 |
| 558 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
-0.666 |
1.383 |
| 559 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.664 |
0.123 |
| 560 |
252412_at |
AT3G47295
|
unknown |
-0.663 |
0.131 |
| 561 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
-0.662 |
0.531 |
| 562 |
249482_at |
AT5G38980
|
unknown |
-0.661 |
0.800 |
| 563 |
253244_at |
AT4G34580
|
AtSFH1, COW1, CAN OF WORMS1, SRH1, SHORT ROOT HAIR 1 |
-0.657 |
0.258 |
| 564 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.655 |
-0.286 |
| 565 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
-0.654 |
-0.021 |
| 566 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.650 |
0.716 |
| 567 |
260639_at |
AT1G53180
|
unknown |
-0.650 |
0.732 |
| 568 |
257173_at |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
-0.648 |
1.873 |
| 569 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.647 |
0.786 |
| 570 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.647 |
0.437 |
| 571 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.646 |
1.195 |
| 572 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-0.646 |
0.813 |
| 573 |
245906_at |
AT5G11070
|
unknown |
-0.643 |
0.180 |
| 574 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.642 |
0.900 |
| 575 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.641 |
0.768 |
| 576 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
-0.640 |
-0.033 |
| 577 |
258480_at |
AT3G02640
|
unknown |
-0.639 |
1.196 |
| 578 |
264987_at |
AT1G27030
|
unknown |
-0.637 |
1.206 |
| 579 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.636 |
0.142 |
| 580 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.636 |
0.388 |
| 581 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
-0.634 |
1.416 |
| 582 |
246270_at |
AT4G36500
|
unknown |
-0.634 |
-0.058 |
| 583 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.631 |
1.225 |
| 584 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.631 |
0.394 |
| 585 |
248718_at |
AT5G47770
|
FPS1, farnesyl diphosphate synthase 1 |
-0.629 |
0.799 |
| 586 |
267287_at |
AT2G23630
|
sks16, SKU5 similar 16 |
-0.628 |
0.664 |
| 587 |
246487_at |
AT5G16030
|
unknown |
-0.627 |
1.006 |
| 588 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.625 |
-0.205 |
| 589 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.625 |
-0.100 |
| 590 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.624 |
0.886 |
| 591 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.623 |
1.047 |
| 592 |
262373_at |
AT1G73120
|
unknown |
-0.622 |
0.696 |
| 593 |
246505_at |
AT5G16250
|
unknown |
-0.622 |
0.713 |
| 594 |
260387_at |
AT1G74100
|
ATSOT16, SULFOTRANSFERASE 16, ATST5A, ARABIDOPSIS SULFOTRANSFERASE 5A, CORI-7, CORONATINE INDUCED-7, SOT16, sulfotransferase 16 |
-0.621 |
0.844 |
| 595 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
-0.620 |
0.466 |
| 596 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.620 |
0.807 |
| 597 |
250936_at |
AT5G03120
|
unknown |
-0.619 |
-0.058 |
| 598 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
-0.619 |
0.566 |
| 599 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.617 |
0.560 |
| 600 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.616 |
1.173 |