|
probeID |
AGICode |
Annotation |
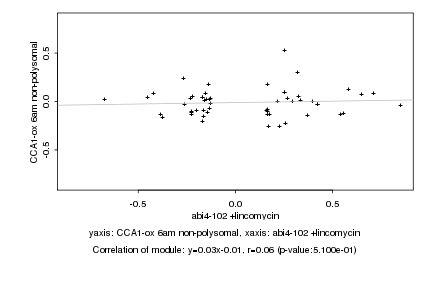
Log2 signal ratio
abi4-102 +lincomycin |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.847 |
-0.031 |
| 2 |
250979_at |
AT5G03090
|
unknown |
0.710 |
0.086 |
| 3 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.648 |
0.075 |
| 4 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.578 |
0.134 |
| 5 |
249114_at |
AT5G43800
|
[copia-like retrotransposon family, has a 0. P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
0.553 |
-0.117 |
| 6 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.537 |
-0.129 |
| 7 |
245766_at |
AT1G33580
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 5.5e-96 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.420 |
-0.028 |
| 8 |
263089_at |
AT2G16000
|
[copia-like retrotransposon family, has a 2.6e-248 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
0.417 |
-0.022 |
| 9 |
263346_at |
AT2G05660
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.392 |
0.009 |
| 10 |
257551_at |
AT3G13270
|
unknown |
0.366 |
-0.143 |
| 11 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
0.330 |
0.012 |
| 12 |
248509_at |
AT5G50335
|
unknown |
0.323 |
0.055 |
| 13 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
0.317 |
0.302 |
| 14 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.291 |
0.007 |
| 15 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.266 |
0.033 |
| 16 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.253 |
-0.219 |
| 17 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.251 |
0.537 |
| 18 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.250 |
0.101 |
| 19 |
246888_at |
AT5G26270
|
unknown |
0.249 |
0.099 |
| 20 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
0.222 |
-0.257 |
| 21 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.212 |
0.002 |
| 22 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.175 |
-0.127 |
| 23 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.165 |
-0.258 |
| 24 |
252865_at |
AT4G39753
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.163 |
-0.101 |
| 25 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
0.162 |
0.186 |
| 26 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.162 |
-0.128 |
| 27 |
266321_at |
AT2G46660
|
CYP78A6, cytochrome P450, family 78, subfamily A, polypeptide 6, EOD3, enhancer of da1-1 |
0.161 |
-0.079 |
| 28 |
257284_at |
AT3G29650
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 5.1e-81 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
0.157 |
-0.083 |
| 29 |
257144_at |
AT3G27300
|
G6PD5, glucose-6-phosphate dehydrogenase 5 |
-0.678 |
0.024 |
| 30 |
248549_at |
AT5G50310
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.457 |
0.044 |
| 31 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
-0.424 |
0.084 |
| 32 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.387 |
-0.125 |
| 33 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.379 |
-0.161 |
| 34 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
-0.272 |
0.244 |
| 35 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.264 |
-0.027 |
| 36 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.235 |
0.033 |
| 37 |
263826_at |
AT2G40410
|
AtCaN2, Ca2+-dependent nuclease, CAN2, calcium dependent nuclease 2 |
-0.231 |
-0.129 |
| 38 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.230 |
-0.098 |
| 39 |
261441_at |
AT1G28470
|
ANAC010, NAC domain containing protein 10, NAC010, NAC domain containing protein 10, SND3, SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 3 |
-0.229 |
-0.111 |
| 40 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-0.226 |
0.057 |
| 41 |
263387_at |
AT2G40160
|
TBL30 |
-0.202 |
-0.083 |
| 42 |
262261_at |
AT1G70895
|
CLE17, CLAVATA3/ESR-RELATED 17 |
-0.174 |
-0.205 |
| 43 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
-0.174 |
0.046 |
| 44 |
246200_at |
AT4G37240
|
unknown |
-0.170 |
0.045 |
| 45 |
267344_at |
AT2G44230
|
unknown |
-0.168 |
-0.149 |
| 46 |
258438_at |
AT3G17230
|
[invertase/pectin methylesterase inhibitor family protein] |
-0.168 |
-0.086 |
| 47 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
-0.160 |
0.012 |
| 48 |
264528_at |
AT1G30810
|
JMJ18, Jumonji domain-containing protein 18 |
-0.156 |
0.083 |
| 49 |
253930_at |
AT4G26740
|
AtCLO1, ATPXG1, ARABIDOPSIS THALIANA PEROXYGENASE 1, ATS1, seed gene 1, CLO1, CALEOSIN1 |
-0.153 |
0.031 |
| 50 |
251612_at |
AT3G57950
|
unknown |
-0.145 |
-0.111 |
| 51 |
260128_at |
AT1G36310
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.139 |
0.182 |
| 52 |
254123_at |
AT4G24640
|
APPB1 |
-0.138 |
0.022 |
| 53 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
-0.138 |
-0.063 |
| 54 |
263105_at |
AT2G05320
|
[beta-1,2-N-acetylglucosaminyltransferase II] |
-0.133 |
-0.016 |
| 55 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.132 |
0.032 |