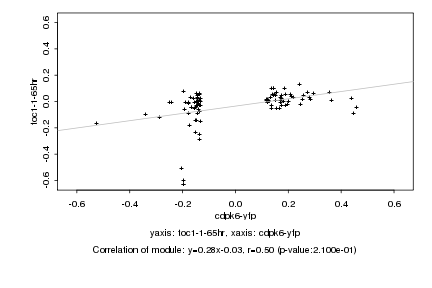
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cdpk6-yfp |
Log2 signal ratio
toc1-1-65hr |
| 1 |
263515_at |
AT2G21640
|
unknown |
0.458 |
-0.043 |
| 2 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.446 |
-0.088 |
| 3 |
257817_at |
AT3G25150
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.437 |
0.027 |
| 4 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.363 |
0.008 |
| 5 |
260522_x_at |
AT2G41730
|
unknown |
0.354 |
0.070 |
| 6 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.294 |
0.063 |
| 7 |
260367_at |
AT1G69760
|
unknown |
0.281 |
0.021 |
| 8 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.277 |
0.038 |
| 9 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.270 |
0.070 |
| 10 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.257 |
0.046 |
| 11 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.250 |
0.019 |
| 12 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.245 |
-0.018 |
| 13 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.240 |
0.135 |
| 14 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.218 |
0.031 |
| 15 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.211 |
0.043 |
| 16 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.207 |
0.058 |
| 17 |
258074_at |
AT3G25890
|
CRF11, cytokinin response factor 11 |
0.200 |
0.005 |
| 18 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.196 |
-0.019 |
| 19 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.189 |
0.054 |
| 20 |
262719_at |
AT1G43590
|
unknown |
0.188 |
-0.023 |
| 21 |
255111_at |
AT4G08780
|
[Peroxidase superfamily protein] |
0.182 |
0.105 |
| 22 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.181 |
0.005 |
| 23 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.174 |
0.029 |
| 24 |
266574_at |
AT2G23890
|
[HAD-superfamily hydrolase, subfamily IG, 5'-nucleotidase] |
0.174 |
-0.024 |
| 25 |
254238_at |
AT4G23540
|
[ARM repeat superfamily protein] |
0.172 |
0.048 |
| 26 |
266471_at |
AT2G31060
|
EMB2785, EMBRYO DEFECTIVE 2785 |
0.170 |
0.031 |
| 27 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.170 |
0.000 |
| 28 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.167 |
0.012 |
| 29 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.166 |
-0.053 |
| 30 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.153 |
-0.053 |
| 31 |
252754_at |
AT3G43510
|
[copia-like retrotransposon family, has a 2.3e-11 P-value blast match to GB:BAA11674 ORF(AA 1-1338) (Ty1_Copia-element) (Nicotiana tabacum)] |
0.153 |
0.075 |
| 32 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.151 |
0.015 |
| 33 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.151 |
0.052 |
| 34 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.147 |
0.058 |
| 35 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.142 |
0.102 |
| 36 |
263061_at |
AT2G18190
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.141 |
0.100 |
| 37 |
259227_at |
AT3G07750
|
[3'-5'-exoribonuclease family protein] |
0.141 |
0.053 |
| 38 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
0.139 |
0.057 |
| 39 |
249702_at |
AT5G35570
|
[O-fucosyltransferase family protein] |
0.136 |
-0.026 |
| 40 |
258996_at |
AT3G01800
|
[Ribosome recycling factor] |
0.135 |
0.099 |
| 41 |
255279_at |
AT4G04950
|
AtGRXS17, Arabidopsis thaliana monothiol glutaredoxin 17, GRXS17, monothiol glutaredoxin 17 |
0.133 |
-0.050 |
| 42 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
0.132 |
0.031 |
| 43 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
0.124 |
0.003 |
| 44 |
265850_at |
AT2G35720
|
OWL1, ORIENTATION UNDER VERY LOW FLUENCES OF LIGHT 1 |
0.122 |
0.015 |
| 45 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.121 |
0.000 |
| 46 |
258131_at |
AT3G24540
|
AtPERK3, proline-rich extensin-like receptor kinase 3, PERK3, proline-rich extensin-like receptor kinase 3 |
0.120 |
-0.001 |
| 47 |
250689_at |
AT5G06610
|
unknown |
0.119 |
-0.004 |
| 48 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.118 |
0.019 |
| 49 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
0.117 |
0.014 |
| 50 |
245164_at |
AT2G33210
|
HSP60-2, heat shock protein 60-2 |
0.117 |
0.016 |
| 51 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.528 |
-0.165 |
| 52 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.343 |
-0.098 |
| 53 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.288 |
-0.115 |
| 54 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.252 |
-0.007 |
| 55 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.245 |
-0.003 |
| 56 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.206 |
-0.506 |
| 57 |
250778_at |
AT5G05500
|
MOP10 |
-0.200 |
-0.623 |
| 58 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.200 |
0.078 |
| 59 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.200 |
-0.623 |
| 60 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.199 |
-0.595 |
| 61 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.196 |
-0.060 |
| 62 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
-0.192 |
-0.003 |
| 63 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
-0.180 |
-0.011 |
| 64 |
250664_at |
AT5G07080
|
[HXXXD-type acyl-transferase family protein] |
-0.180 |
-0.002 |
| 65 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
-0.178 |
-0.087 |
| 66 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.174 |
-0.178 |
| 67 |
257067_at |
AT3G18270
|
pseudogene |
-0.173 |
0.032 |
| 68 |
253527_at |
AT4G31470
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.169 |
-0.045 |
| 69 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
-0.167 |
-0.045 |
| 70 |
262595_at |
AT1G15360
|
AtSHN1, SHN1, SHINE 1, WIN1, WAX INDUCER 1 |
-0.162 |
0.023 |
| 71 |
261026_at |
AT1G01240
|
unknown |
-0.157 |
-0.004 |
| 72 |
263699_at |
AT1G31120
|
KUP10, K+ uptake permease 10 |
-0.157 |
-0.049 |
| 73 |
265740_at |
AT2G01150
|
RHA2B, RING-H2 finger protein 2B |
-0.154 |
-0.144 |
| 74 |
256671_at |
AT3G52290
|
IQD3, IQ-domain 3 |
-0.154 |
-0.039 |
| 75 |
257761_at |
AT3G23090
|
WDL3 |
-0.152 |
-0.235 |
| 76 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
-0.151 |
-0.025 |
| 77 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.148 |
-0.141 |
| 78 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.148 |
0.064 |
| 79 |
256599_at |
AT3G14760
|
unknown |
-0.148 |
0.007 |
| 80 |
250881_at |
AT5G04080
|
unknown |
-0.147 |
-0.087 |
| 81 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.147 |
0.029 |
| 82 |
253454_at |
AT4G31875
|
unknown |
-0.145 |
0.053 |
| 83 |
254263_at |
AT4G23493
|
unknown |
-0.145 |
0.005 |
| 84 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.144 |
0.033 |
| 85 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.144 |
-0.028 |
| 86 |
245792_at |
AT1G32100
|
ATPRR1, PRR1, pinoresinol reductase 1 |
-0.143 |
-0.054 |
| 87 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.143 |
0.001 |
| 88 |
251394_at |
AT3G60900
|
FLA10, FASCICLIN-like arabinogalactan-protein 10 |
-0.142 |
-0.013 |
| 89 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.139 |
0.012 |
| 90 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.138 |
-0.249 |
| 91 |
247965_at |
AT5G56540
|
AGP14, arabinogalactan protein 14, ATAGP14 |
-0.137 |
-0.288 |
| 92 |
262313_at |
AT1G70900
|
unknown |
-0.137 |
-0.070 |
| 93 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.137 |
0.066 |
| 94 |
253111_at |
AT4G35940
|
unknown |
-0.136 |
0.057 |
| 95 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
-0.136 |
-0.025 |
| 96 |
264539_at |
AT1G55590
|
[RNI-like superfamily protein] |
-0.134 |
0.001 |
| 97 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
-0.134 |
-0.146 |
| 98 |
265237_s_at |
ATMG00470
|
ORF122A |
-0.133 |
0.027 |
| 99 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
-0.133 |
-0.023 |