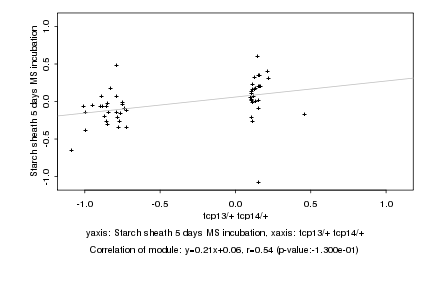
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
tcp13/+ tcp14/+ |
Log2 signal ratio
Starch sheath 5 days MS incubation |
| 1 |
248545_at |
AT5G50260
|
CEP1, cysteine endopeptidase 1 |
0.452 |
-0.172 |
| 2 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.216 |
0.311 |
| 3 |
245993_at |
AT5G20700
|
unknown |
0.207 |
0.405 |
| 4 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.162 |
0.208 |
| 5 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
0.158 |
0.357 |
| 6 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.156 |
0.209 |
| 7 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.152 |
0.348 |
| 8 |
266530_at |
AT2G16910
|
AMS, ABORTED MICROSPORES |
0.150 |
0.019 |
| 9 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
0.150 |
-0.081 |
| 10 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
0.150 |
-1.079 |
| 11 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.147 |
0.208 |
| 12 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.144 |
0.606 |
| 13 |
260690_at |
AT1G32340
|
NHL8, NDR1/HIN1-like 8 |
0.132 |
0.185 |
| 14 |
255707_at |
AT4G00231
|
MEE50, maternal effect embryo arrest 50 |
0.129 |
0.013 |
| 15 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
0.122 |
0.331 |
| 16 |
246567_at |
AT5G14950
|
ATGMII, GMII, golgi alpha-mannosidase II |
0.121 |
0.169 |
| 17 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.115 |
0.073 |
| 18 |
264801_at |
AT1G08840
|
emb2411, embryo defective 2411 |
0.113 |
0.166 |
| 19 |
267549_at |
AT2G32640
|
[Lycopene beta/epsilon cyclase protein] |
0.112 |
-0.013 |
| 20 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.108 |
0.237 |
| 21 |
251530_at |
AT3G58520
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.107 |
-0.259 |
| 22 |
261505_at |
AT1G71696
|
SOL1, SUPPRESSOR OF LLP1 1 |
0.105 |
0.024 |
| 23 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
0.104 |
-0.207 |
| 24 |
252514_at |
AT3G46060
|
ARA-3, ARA3, ATRAB8A, RAB GTPase homolog 8A, ATRABE1C, RAB8A, RAB GTPase homolog 8A, RABE1c |
0.104 |
0.109 |
| 25 |
250419_at |
AT5G11250
|
[Disease resistance protein (TIR-NBS-LRR class)] |
0.103 |
0.140 |
| 26 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
0.103 |
0.036 |
| 27 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
0.099 |
0.063 |
| 28 |
259266_at |
AT3G01240
|
unknown |
-1.093 |
-0.641 |
| 29 |
248714_at |
AT5G48140
|
[Pectin lyase-like superfamily protein] |
-1.009 |
-0.062 |
| 30 |
248470_at |
AT5G50830
|
unknown |
-1.001 |
-0.141 |
| 31 |
249429_at |
AT5G39880
|
unknown |
-0.996 |
-0.378 |
| 32 |
253831_at |
AT4G27580
|
unknown |
-0.951 |
-0.046 |
| 33 |
257819_at |
AT3G25165
|
RALFL25, ralf-like 25 |
-0.901 |
-0.063 |
| 34 |
246545_at |
AT5G15110
|
[Pectate lyase family protein] |
-0.892 |
0.075 |
| 35 |
257821_at |
AT3G25170
|
RALFL26, ralf-like 26 |
-0.889 |
-0.054 |
| 36 |
261528_at |
AT1G14420
|
AT59 |
-0.870 |
-0.194 |
| 37 |
256945_at |
AT3G19020
|
[Leucine-rich repeat (LRR) family protein] |
-0.858 |
-0.055 |
| 38 |
261623_at |
AT1G01980
|
[ATSEC1A] |
-0.857 |
-0.262 |
| 39 |
262022_at |
AT1G35490
|
[bZIP family transcription factor] |
-0.853 |
-0.023 |
| 40 |
248194_at |
AT5G54095
|
unknown |
-0.851 |
-0.293 |
| 41 |
248822_at |
AT5G47000
|
[Peroxidase superfamily protein] |
-0.845 |
-0.138 |
| 42 |
267476_at |
AT2G02720
|
[Pectate lyase family protein] |
-0.831 |
0.178 |
| 43 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
-0.795 |
0.486 |
| 44 |
262664_at |
AT1G13970
|
unknown |
-0.793 |
-0.146 |
| 45 |
251854_at |
AT3G54800
|
[Pleckstrin homology (PH) and lipid-binding START domains-containing protein] |
-0.791 |
0.073 |
| 46 |
250631_at |
AT5G07430
|
[Pectin lyase-like superfamily protein] |
-0.784 |
-0.202 |
| 47 |
251252_at |
AT3G62230
|
DAF1, DUO1-activated F-box 1 |
-0.777 |
-0.345 |
| 48 |
251180_at |
AT3G62640
|
unknown |
-0.772 |
-0.260 |
| 49 |
265368_at |
AT2G13350
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.764 |
-0.155 |
| 50 |
263450_at |
AT2G31500
|
CPK24, calcium-dependent protein kinase 24 |
-0.756 |
-0.036 |
| 51 |
266918_at |
AT2G45800
|
PLIM2a, PLIM2a |
-0.750 |
-0.003 |
| 52 |
257090_at |
AT3G20530
|
[Protein kinase superfamily protein] |
-0.738 |
-0.086 |
| 53 |
262696_at |
AT1G75870
|
unknown |
-0.727 |
-0.337 |
| 54 |
245700_at |
AT5G04180
|
ACA3, alpha carbonic anhydrase 3, ATACA3, ALPHA CARBONIC ANHYDRASE 3 |
-0.724 |
-0.115 |