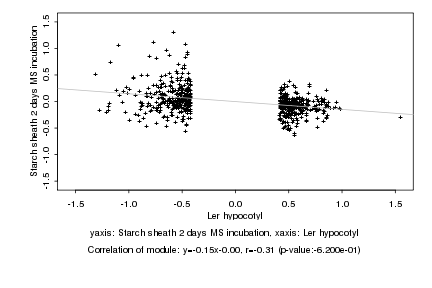
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ler hypocotyl |
Log2 signal ratio
Starch sheath 2 days MS incubation |
| 1 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
1.545 |
-0.296 |
| 2 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
0.984 |
-0.148 |
| 3 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.968 |
-0.116 |
| 4 |
246080_at |
AT5G20460
|
unknown |
0.946 |
-0.012 |
| 5 |
263081_at |
AT2G27220
|
BLH5, BEL1-like homeodomain 5 |
0.932 |
-0.105 |
| 6 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
0.912 |
-0.119 |
| 7 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.884 |
-0.121 |
| 8 |
245454_at |
AT4G16920
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.870 |
-0.008 |
| 9 |
262421_at |
AT1G50290
|
unknown |
0.865 |
-0.093 |
| 10 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.858 |
-0.246 |
| 11 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.853 |
-0.062 |
| 12 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.853 |
-0.290 |
| 13 |
254566_at |
AT4G19240
|
unknown |
0.851 |
-0.047 |
| 14 |
261221_at |
AT1G19960
|
unknown |
0.851 |
-0.317 |
| 15 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.846 |
0.212 |
| 16 |
267414_at |
AT2G34790
|
EDA28, EMBRYO SAC DEVELOPMENT ARREST 28, MEE23, MATERNAL EFFECT EMBRYO ARREST 23 |
0.842 |
-0.102 |
| 17 |
253532_at |
AT4G31570
|
unknown |
0.829 |
0.041 |
| 18 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.829 |
-0.015 |
| 19 |
252345_at |
AT3G48640
|
unknown |
0.829 |
-0.178 |
| 20 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.824 |
-0.358 |
| 21 |
246967_at |
AT5G24860
|
ATFPF1, ARABIDOPSIS FLOWERING PROMOTING FACTOR 1, FPF1, FLOWERING PROMOTING FACTOR 1 |
0.824 |
-0.207 |
| 22 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
0.822 |
-0.155 |
| 23 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
0.813 |
0.016 |
| 24 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.809 |
0.037 |
| 25 |
262903_at |
AT1G59950
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.807 |
-0.050 |
| 26 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.802 |
0.001 |
| 27 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.799 |
0.061 |
| 28 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.788 |
-0.139 |
| 29 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
0.786 |
-0.051 |
| 30 |
250135_at |
AT5G15360
|
unknown |
0.778 |
-0.168 |
| 31 |
254574_at |
AT4G19430
|
unknown |
0.777 |
0.006 |
| 32 |
257417_at |
AT1G10110
|
[F-box family protein] |
0.767 |
0.056 |
| 33 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.767 |
-0.475 |
| 34 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.764 |
-0.312 |
| 35 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
0.761 |
-0.152 |
| 36 |
247751_at |
AT5G59050
|
unknown |
0.758 |
0.095 |
| 37 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.758 |
-0.065 |
| 38 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.754 |
-0.178 |
| 39 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.753 |
-0.103 |
| 40 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.747 |
-0.144 |
| 41 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
0.746 |
-0.137 |
| 42 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
0.742 |
-0.126 |
| 43 |
251197_at |
AT3G62960
|
[Thioredoxin superfamily protein] |
0.741 |
-0.089 |
| 44 |
253827_at |
AT4G28085
|
unknown |
0.740 |
-0.117 |
| 45 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.724 |
0.017 |
| 46 |
265998_at |
AT2G24270
|
ALDH11A3, aldehyde dehydrogenase 11A3 |
0.718 |
-0.136 |
| 47 |
256515_at |
AT1G66020
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.701 |
-0.166 |
| 48 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.693 |
0.337 |
| 49 |
266097_at |
AT2G37970
|
AtHBP2, HBP2, haem-binding protein 2, SOUL-1 |
0.691 |
0.285 |
| 50 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.685 |
0.006 |
| 51 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
0.681 |
0.167 |
| 52 |
264363_at |
AT1G03170
|
FAF2, FANTASTIC FOUR 2, FTM5, FLORAL TRANSITION AT THE MERISTEM5 |
0.679 |
0.032 |
| 53 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.677 |
0.031 |
| 54 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.677 |
-0.064 |
| 55 |
257899_at |
AT3G28345
|
ABCB15, ATP-binding cassette B15, MDR13, multi-drug resistance 13 |
0.673 |
-0.055 |
| 56 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.673 |
-0.050 |
| 57 |
257375_at |
AT2G38640
|
unknown |
0.670 |
-0.044 |
| 58 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
0.667 |
-0.137 |
| 59 |
260639_at |
AT1G53180
|
unknown |
0.666 |
-0.215 |
| 60 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.663 |
-0.252 |
| 61 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.662 |
-0.372 |
| 62 |
254384_at |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
0.662 |
0.051 |
| 63 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.660 |
-0.348 |
| 64 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.660 |
-0.019 |
| 65 |
258807_at |
AT3G04030
|
MYR2 |
0.658 |
-0.085 |
| 66 |
251940_at |
AT3G53450
|
LOG4, LONELY GUY 4 |
0.658 |
-0.032 |
| 67 |
267459_at |
AT2G33850
|
unknown |
0.657 |
-0.104 |
| 68 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.651 |
-0.117 |
| 69 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
0.638 |
0.036 |
| 70 |
247707_at |
AT5G59450
|
[GRAS family transcription factor] |
0.634 |
-0.152 |
| 71 |
258306_at |
AT3G30530
|
ATBZIP42, basic leucine-zipper 42, bZIP42, basic leucine-zipper 42 |
0.634 |
-0.033 |
| 72 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.628 |
-0.072 |
| 73 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
0.625 |
-0.098 |
| 74 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.625 |
0.030 |
| 75 |
266824_at |
AT2G22800
|
HAT9 |
0.624 |
0.060 |
| 76 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.623 |
0.034 |
| 77 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
0.621 |
-0.217 |
| 78 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.620 |
-0.084 |
| 79 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.620 |
-0.181 |
| 80 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.619 |
-0.170 |
| 81 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.617 |
0.054 |
| 82 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.614 |
-0.163 |
| 83 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.613 |
-0.044 |
| 84 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
0.612 |
-0.288 |
| 85 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.611 |
0.008 |
| 86 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.611 |
-0.068 |
| 87 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.610 |
-0.072 |
| 88 |
255945_at |
AT5G28610
|
unknown |
0.608 |
-0.167 |
| 89 |
248563_at |
AT5G49690
|
[UDP-Glycosyltransferase superfamily protein] |
0.604 |
-0.139 |
| 90 |
247051_at |
AT5G66670
|
unknown |
0.603 |
-0.088 |
| 91 |
266995_at |
AT2G34500
|
CYP710A1, cytochrome P450, family 710, subfamily A, polypeptide 1 |
0.603 |
-0.212 |
| 92 |
260640_at |
AT1G53350
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.601 |
-0.053 |
| 93 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.600 |
-0.124 |
| 94 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.599 |
0.044 |
| 95 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.595 |
0.086 |
| 96 |
253243_at |
AT4G34560
|
unknown |
0.595 |
-0.007 |
| 97 |
259661_at |
AT1G55265
|
unknown |
0.587 |
-0.057 |
| 98 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
0.587 |
-0.237 |
| 99 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
0.583 |
-0.041 |
| 100 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
0.583 |
-0.119 |
| 101 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
0.582 |
0.064 |
| 102 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.577 |
-0.027 |
| 103 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.576 |
-0.046 |
| 104 |
265052_at |
AT1G51990
|
[O-methyltransferase family protein] |
0.575 |
-0.350 |
| 105 |
252458_at |
AT3G47210
|
unknown |
0.574 |
-0.398 |
| 106 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.574 |
0.154 |
| 107 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
0.572 |
0.158 |
| 108 |
261684_at |
AT1G47400
|
unknown |
0.572 |
-0.074 |
| 109 |
260472_at |
AT1G10990
|
unknown |
0.572 |
-0.128 |
| 110 |
264562_at |
AT1G55760
|
[BTB/POZ domain-containing protein] |
0.570 |
-0.220 |
| 111 |
259032_at |
AT3G09380
|
unknown |
0.568 |
0.001 |
| 112 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.567 |
-0.464 |
| 113 |
255955_at |
AT1G22030
|
unknown |
0.567 |
-0.311 |
| 114 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.566 |
-0.095 |
| 115 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.566 |
0.060 |
| 116 |
249797_at |
AT5G23750
|
[Remorin family protein] |
0.564 |
-0.110 |
| 117 |
262526_at |
AT1G17050
|
AtSPS2, SPS2, solanesyl diphosphate synthase 2 |
0.560 |
0.273 |
| 118 |
264580_at |
AT1G05340
|
unknown |
0.556 |
-0.072 |
| 119 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.555 |
0.035 |
| 120 |
252383_at |
AT3G47780
|
ABCA7, ATP-binding cassette A7, ATATH6, A. THALIANA ABC2 HOMOLOG 6, ATH6, ABC2 homolog 6 |
0.555 |
-0.085 |
| 121 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.555 |
-0.064 |
| 122 |
254201_at |
AT4G24130
|
unknown |
0.555 |
-0.223 |
| 123 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
0.555 |
0.176 |
| 124 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
0.552 |
-0.584 |
| 125 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.549 |
-0.036 |
| 126 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
0.549 |
-0.252 |
| 127 |
245690_at |
AT5G04230
|
ATPAL3, PAL3, phenyl alanine ammonia-lyase 3 |
0.548 |
-0.199 |
| 128 |
262286_at |
AT1G68585
|
unknown |
0.548 |
-0.258 |
| 129 |
259430_at |
AT1G01610
|
ATGPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4, GPAT4, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 4 |
0.547 |
-0.007 |
| 130 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
0.545 |
0.041 |
| 131 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.545 |
0.048 |
| 132 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
0.545 |
-0.638 |
| 133 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.544 |
-0.351 |
| 134 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.544 |
0.000 |
| 135 |
260975_at |
AT1G53430
|
[Leucine-rich repeat transmembrane protein kinase] |
0.541 |
-0.101 |
| 136 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.540 |
-0.127 |
| 137 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
0.540 |
-0.013 |
| 138 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
0.538 |
-0.079 |
| 139 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.537 |
-0.144 |
| 140 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
0.537 |
0.313 |
| 141 |
247035_at |
AT5G67110
|
ALC, ALCATRAZ |
0.537 |
0.055 |
| 142 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
0.536 |
-0.092 |
| 143 |
252661_at |
AT3G44450
|
unknown |
0.534 |
-0.221 |
| 144 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.534 |
-0.076 |
| 145 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.532 |
-0.024 |
| 146 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
0.532 |
-0.385 |
| 147 |
253024_at |
AT4G38080
|
[hydroxyproline-rich glycoprotein family protein] |
0.530 |
-0.041 |
| 148 |
264151_at |
AT1G02070
|
unknown |
0.528 |
-0.139 |
| 149 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
0.526 |
0.073 |
| 150 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.526 |
0.016 |
| 151 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.525 |
-0.187 |
| 152 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.524 |
-0.005 |
| 153 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.522 |
-0.019 |
| 154 |
256443_at |
AT3G10960
|
ATAZG1, AZA-guanine resistant1, AZG1, AZA-guanine resistant1 |
0.521 |
0.000 |
| 155 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
0.519 |
0.010 |
| 156 |
253525_at |
AT4G31330
|
unknown |
0.519 |
-0.131 |
| 157 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
0.518 |
-0.312 |
| 158 |
245305_at |
AT4G17215
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.516 |
-0.228 |
| 159 |
245558_at |
AT4G15430
|
[ERD (early-responsive to dehydration stress) family protein] |
0.514 |
-0.496 |
| 160 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.514 |
-0.263 |
| 161 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
0.510 |
-0.091 |
| 162 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.508 |
-0.035 |
| 163 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.506 |
-0.530 |
| 164 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.506 |
-0.068 |
| 165 |
257490_x_at |
AT1G01380
|
ETC1, ENHANCER OF TRY AND CPC 1 |
0.505 |
0.047 |
| 166 |
248695_at |
AT5G48350
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.504 |
-0.118 |
| 167 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.504 |
-0.188 |
| 168 |
266129_at |
AT2G44990
|
ATCCD7, CCD7, carotenoid cleavage dioxygenase 7, MAX3 |
0.503 |
-0.067 |
| 169 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.503 |
-0.145 |
| 170 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.503 |
0.387 |
| 171 |
254185_at |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
0.501 |
-0.030 |
| 172 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
0.500 |
0.013 |
| 173 |
262304_at |
AT1G70890
|
MLP43, MLP-like protein 43 |
0.500 |
-0.153 |
| 174 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
0.499 |
-0.090 |
| 175 |
247727_at |
AT5G59490
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.498 |
-0.089 |
| 176 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
0.495 |
-0.291 |
| 177 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.495 |
-0.049 |
| 178 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
0.494 |
-0.435 |
| 179 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
0.494 |
-0.057 |
| 180 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
0.494 |
-0.187 |
| 181 |
253193_at |
AT4G35380
|
[SEC7-like guanine nucleotide exchange family protein] |
0.494 |
-0.490 |
| 182 |
251009_at |
AT5G02640
|
unknown |
0.491 |
-0.097 |
| 183 |
262811_at |
AT1G11700
|
unknown |
0.491 |
-0.209 |
| 184 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.490 |
-0.250 |
| 185 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.489 |
-0.113 |
| 186 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.489 |
-0.156 |
| 187 |
254571_at |
AT4G19370
|
unknown |
0.488 |
-0.002 |
| 188 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.487 |
0.296 |
| 189 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.487 |
-0.260 |
| 190 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
0.486 |
0.150 |
| 191 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.484 |
0.062 |
| 192 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
0.483 |
-0.389 |
| 193 |
246270_at |
AT4G36500
|
unknown |
0.483 |
-0.527 |
| 194 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
0.483 |
-0.270 |
| 195 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.482 |
0.066 |
| 196 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.482 |
-0.084 |
| 197 |
262303_at |
AT1G70920
|
ATHB18, homeobox-leucine zipper protein 18, HB18, homeobox-leucine zipper protein 18 |
0.479 |
-0.297 |
| 198 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.479 |
-0.002 |
| 199 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
0.478 |
-0.038 |
| 200 |
257348_at |
AT2G42140
|
[VQ motif-containing protein] |
0.478 |
-0.037 |
| 201 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.477 |
0.023 |
| 202 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.477 |
-0.033 |
| 203 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.475 |
-0.213 |
| 204 |
249794_at |
AT5G23530
|
AtCXE18, carboxyesterase 18, CXE18, carboxyesterase 18 |
0.475 |
-0.078 |
| 205 |
263407_at |
AT2G04038
|
AtbZIP48, basic leucine-zipper 48, bZIP48, basic leucine-zipper 48 |
0.473 |
-0.031 |
| 206 |
250498_at |
AT5G09660
|
PMDH2, peroxisomal NAD-malate dehydrogenase 2 |
0.472 |
0.123 |
| 207 |
259952_at |
AT1G71400
|
AtRLP12, receptor like protein 12, RLP12, receptor like protein 12 |
0.471 |
0.107 |
| 208 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.470 |
-0.013 |
| 209 |
260005_at |
AT1G67920
|
unknown |
0.468 |
-0.385 |
| 210 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
0.468 |
-0.029 |
| 211 |
248801_at |
AT5G47370
|
HAT2 |
0.467 |
-0.463 |
| 212 |
252233_at |
AT3G49690
|
ATMYB84, MYB DOMAIN PROTEIN 84, MYB84, myb domain protein 84, RAX3, REGULATOR OF AXILLARY MERISTEMS3 |
0.467 |
0.098 |
| 213 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.467 |
0.153 |
| 214 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.467 |
-0.074 |
| 215 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.467 |
-0.081 |
| 216 |
266287_at |
AT2G29150
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.466 |
-0.065 |
| 217 |
266396_at |
AT2G38790
|
unknown |
0.466 |
-0.286 |
| 218 |
247359_at |
AT5G63560
|
FACT, FATTY ALCOHOL:CAFFEOYL-CoA CAFFEOYL TRANSFERASE |
0.465 |
-0.069 |
| 219 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
0.464 |
0.048 |
| 220 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
0.464 |
-0.269 |
| 221 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.464 |
-0.298 |
| 222 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.463 |
-0.080 |
| 223 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
0.462 |
-0.138 |
| 224 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.461 |
-0.108 |
| 225 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
0.459 |
-0.103 |
| 226 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
0.459 |
-0.256 |
| 227 |
261570_at |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
0.459 |
-0.080 |
| 228 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.459 |
-0.101 |
| 229 |
267391_at |
AT2G44480
|
BGLU17, beta glucosidase 17 |
0.459 |
0.268 |
| 230 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.458 |
-0.030 |
| 231 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.457 |
0.119 |
| 232 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.457 |
-0.092 |
| 233 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.456 |
0.332 |
| 234 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
0.456 |
-0.011 |
| 235 |
245662_at |
AT1G28190
|
unknown |
0.455 |
0.098 |
| 236 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
0.454 |
-0.245 |
| 237 |
266669_at |
AT2G29750
|
UGT71C1, UDP-glucosyl transferase 71C1 |
0.454 |
-0.260 |
| 238 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.453 |
0.064 |
| 239 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.452 |
-0.052 |
| 240 |
267178_at |
AT2G37750
|
unknown |
0.452 |
-0.091 |
| 241 |
248734_at |
AT5G48090
|
ELP1, EDM2-like protein1 |
0.452 |
0.001 |
| 242 |
264040_at |
AT2G03730
|
ACR5, ACT domain repeat 5 |
0.451 |
-0.477 |
| 243 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
0.451 |
-0.272 |
| 244 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.450 |
-0.200 |
| 245 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.448 |
0.016 |
| 246 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
0.447 |
-0.197 |
| 247 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
0.446 |
0.131 |
| 248 |
255903_at |
AT1G17950
|
ATMYB52, MYB DOMAIN PROTEIN 52, BW52, MYB52, myb domain protein 52 |
0.446 |
-0.501 |
| 249 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.446 |
-0.358 |
| 250 |
266222_at |
AT2G28780
|
unknown |
0.446 |
-0.064 |
| 251 |
262649_at |
AT1G14040
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.443 |
0.227 |
| 252 |
245884_at |
AT5G09300
|
[Thiamin diphosphate-binding fold (THDP-binding) superfamily protein] |
0.442 |
0.093 |
| 253 |
259562_at |
AT1G21200
|
[sequence-specific DNA binding transcription factors] |
0.441 |
-0.039 |
| 254 |
249485_at |
AT5G39020
|
[Malectin/receptor-like protein kinase family protein] |
0.441 |
0.073 |
| 255 |
245090_at |
AT2G40900
|
UMAMIT11, Usually multiple acids move in and out Transporters 11 |
0.441 |
-0.131 |
| 256 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.440 |
0.108 |
| 257 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
0.440 |
-0.059 |
| 258 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.439 |
-0.041 |
| 259 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.438 |
-0.025 |
| 260 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.438 |
-0.185 |
| 261 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
0.438 |
-0.173 |
| 262 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
0.438 |
0.220 |
| 263 |
249987_at |
AT5G18490
|
unknown |
0.437 |
-0.139 |
| 264 |
258275_at |
AT3G15760
|
unknown |
0.435 |
-0.056 |
| 265 |
249134_at |
AT5G43150
|
unknown |
0.433 |
-0.407 |
| 266 |
245740_at |
AT1G44100
|
AAP5, amino acid permease 5 |
0.431 |
0.227 |
| 267 |
262772_at |
AT1G13210
|
ACA.l, autoinhibited Ca2+/ATPase II |
0.431 |
-0.099 |
| 268 |
245352_at |
AT4G15490
|
UGT84A3 |
0.430 |
-0.189 |
| 269 |
265935_at |
AT2G19580
|
TET2, tetraspanin2 |
0.429 |
0.164 |
| 270 |
260696_at |
AT1G32520
|
unknown |
0.428 |
0.016 |
| 271 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
0.428 |
-0.000 |
| 272 |
245702_at |
AT5G04220
|
ATSYTC, NTMC2T1.3, NTMC2TYPE1.3, SYT3, synaptotagmin 3, SYTC |
0.427 |
-0.111 |
| 273 |
253779_at |
AT4G28490
|
HAE, HAESA, RLK5, RECEPTOR-LIKE PROTEIN KINASE 5 |
0.426 |
-0.050 |
| 274 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
0.423 |
0.267 |
| 275 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
0.423 |
-0.115 |
| 276 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
0.422 |
0.156 |
| 277 |
262396_at |
AT1G49470
|
unknown |
0.421 |
0.068 |
| 278 |
262842_at |
AT1G14720
|
ATXTH28, EXGT-A2, ENDOXYLOGLUCAN TRANSFERASE A2, XTH28, xyloglucan endotransglucosylase/hydrolase 28, XTR2, xyloglucan endotransglycosylase related 2 |
0.420 |
0.215 |
| 279 |
257053_at |
AT3G15210
|
ATERF-4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ATERF4, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 4, ERF4, ethylene responsive element binding factor 4, RAP2.5, RELATED TO AP2 5 |
0.420 |
-0.127 |
| 280 |
254408_at |
AT4G21390
|
B120 |
0.420 |
0.079 |
| 281 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.420 |
0.028 |
| 282 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.418 |
-0.064 |
| 283 |
259103_at |
AT3G11690
|
unknown |
0.418 |
-0.103 |
| 284 |
246815_at |
AT5G27220
|
[Frigida-like protein] |
0.418 |
-0.019 |
| 285 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
0.418 |
-0.303 |
| 286 |
255957_at |
AT1G22160
|
unknown |
0.417 |
-0.302 |
| 287 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.417 |
0.033 |
| 288 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.417 |
0.038 |
| 289 |
254256_at |
AT4G23180
|
CRK10, cysteine-rich RLK (RECEPTOR-like protein kinase) 10, RLK4 |
0.416 |
0.035 |
| 290 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
0.416 |
-0.353 |
| 291 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
0.415 |
0.009 |
| 292 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.415 |
-0.090 |
| 293 |
255471_at |
AT4G03050
|
AOP3 |
0.415 |
-0.321 |
| 294 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.415 |
0.017 |
| 295 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.415 |
-0.035 |
| 296 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
0.415 |
-0.215 |
| 297 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
0.414 |
0.221 |
| 298 |
248040_at |
AT5G55970
|
[RING/U-box superfamily protein] |
0.414 |
-0.207 |
| 299 |
259520_at |
AT1G12320
|
unknown |
0.413 |
-0.338 |
| 300 |
253053_at |
AT4G37470
|
KAI2, KARRIKIN INSENSITIVE 2 |
0.413 |
-0.262 |
| 301 |
262206_at |
AT2G01090
|
[Ubiquinol-cytochrome C reductase hinge protein] |
-1.322 |
0.527 |
| 302 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-1.276 |
-0.154 |
| 303 |
245531_at |
AT4G15100
|
scpl30, serine carboxypeptidase-like 30 |
-1.213 |
-0.205 |
| 304 |
252882_at |
AT4G39675
|
unknown |
-1.196 |
-0.089 |
| 305 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-1.193 |
-0.162 |
| 306 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-1.183 |
-0.031 |
| 307 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.178 |
0.749 |
| 308 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
-1.124 |
0.216 |
| 309 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-1.101 |
1.065 |
| 310 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
-1.090 |
0.125 |
| 311 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
-1.060 |
-0.023 |
| 312 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-1.058 |
0.204 |
| 313 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-1.033 |
-0.197 |
| 314 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
-1.025 |
0.280 |
| 315 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
-1.018 |
0.165 |
| 316 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
-1.002 |
0.240 |
| 317 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-1.001 |
-0.344 |
| 318 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.964 |
0.448 |
| 319 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.959 |
0.459 |
| 320 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.946 |
-0.231 |
| 321 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.943 |
0.124 |
| 322 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
-0.917 |
-0.249 |
| 323 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.907 |
0.196 |
| 324 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.903 |
-0.364 |
| 325 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
-0.899 |
-0.009 |
| 326 |
249724_at |
AT5G35450
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.898 |
-0.150 |
| 327 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.886 |
0.496 |
| 328 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.885 |
-0.085 |
| 329 |
261215_at |
AT1G32970
|
[Subtilisin-like serine endopeptidase family protein] |
-0.884 |
-0.029 |
| 330 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.880 |
-0.021 |
| 331 |
248079_at |
AT5G55790
|
unknown |
-0.874 |
-0.068 |
| 332 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.867 |
-0.305 |
| 333 |
258027_at |
AT3G19515
|
unknown |
-0.853 |
-0.208 |
| 334 |
267460_at |
AT2G33810
|
SPL3, squamosa promoter binding protein-like 3 |
-0.839 |
-0.462 |
| 335 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.839 |
0.096 |
| 336 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
-0.837 |
0.162 |
| 337 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.830 |
-0.085 |
| 338 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
-0.826 |
0.101 |
| 339 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
-0.826 |
0.495 |
| 340 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.823 |
-0.031 |
| 341 |
248016_at |
AT5G56380
|
[F-box/RNI-like/FBD-like domains-containing protein] |
-0.822 |
0.262 |
| 342 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.809 |
0.861 |
| 343 |
263544_at |
AT2G21590
|
APL4 |
-0.801 |
0.152 |
| 344 |
261323_at |
AT1G44760
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.799 |
-0.058 |
| 345 |
257322_at |
ATMG01180
|
ORF111B |
-0.798 |
-0.188 |
| 346 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.796 |
0.080 |
| 347 |
256569_at |
AT3G19550
|
unknown |
-0.791 |
-0.160 |
| 348 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-0.774 |
1.126 |
| 349 |
256188_at |
AT1G30160
|
unknown |
-0.772 |
-0.095 |
| 350 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-0.769 |
0.316 |
| 351 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.761 |
0.135 |
| 352 |
263137_at |
AT1G78660
|
ATGGH1, gamma-glutamyl hydrolase 1, GGH1, gamma-glutamyl hydrolase 1 |
-0.750 |
0.184 |
| 353 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.749 |
-0.114 |
| 354 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.747 |
-0.004 |
| 355 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.745 |
-0.156 |
| 356 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
-0.744 |
0.315 |
| 357 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
-0.744 |
-0.412 |
| 358 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
-0.741 |
0.820 |
| 359 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
-0.733 |
-0.151 |
| 360 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
-0.731 |
-0.063 |
| 361 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.728 |
0.057 |
| 362 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.727 |
0.268 |
| 363 |
258228_at |
AT3G27610
|
[Nucleotidylyl transferase superfamily protein] |
-0.726 |
-0.104 |
| 364 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.721 |
-0.190 |
| 365 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
-0.719 |
0.201 |
| 366 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.711 |
0.267 |
| 367 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
-0.710 |
-0.095 |
| 368 |
245341_at |
AT4G16447
|
unknown |
-0.710 |
-0.168 |
| 369 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
-0.709 |
0.398 |
| 370 |
263285_at |
AT2G36120
|
DOT1, DEFECTIVELY ORGANIZED TRIBUTARIES 1 |
-0.708 |
-0.050 |
| 371 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
-0.704 |
0.140 |
| 372 |
252659_at |
AT3G44430
|
unknown |
-0.702 |
-0.028 |
| 373 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
-0.701 |
0.339 |
| 374 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
-0.700 |
0.275 |
| 375 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.700 |
0.092 |
| 376 |
260028_at |
AT1G29980
|
unknown |
-0.696 |
0.474 |
| 377 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.693 |
0.155 |
| 378 |
250611_at |
AT5G07200
|
ATGA20OX3, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 3, GA20OX3, gibberellin 20-oxidase 3, YAP169 |
-0.692 |
-0.011 |
| 379 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.685 |
0.117 |
| 380 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.684 |
-0.005 |
| 381 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
-0.684 |
0.068 |
| 382 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.682 |
-0.284 |
| 383 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-0.681 |
0.044 |
| 384 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
-0.679 |
0.345 |
| 385 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.675 |
-0.027 |
| 386 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.669 |
0.145 |
| 387 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.666 |
-0.264 |
| 388 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.664 |
0.492 |
| 389 |
247666_at |
AT5G60140
|
[AP2/B3-like transcriptional factor family protein] |
-0.663 |
0.018 |
| 390 |
260107_at |
AT1G66430
|
[pfkB-like carbohydrate kinase family protein] |
-0.658 |
0.300 |
| 391 |
250058_at |
AT5G17870
|
PSRP6, plastid-specific 50S ribosomal protein 6 |
-0.657 |
0.132 |
| 392 |
258503_at |
AT3G02500
|
unknown |
-0.654 |
-0.456 |
| 393 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.650 |
0.978 |
| 394 |
259796_at |
AT1G64270
|
[Mutator-like transposase family, has a 1.0e-06 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.645 |
0.007 |
| 395 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.644 |
-0.057 |
| 396 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.643 |
0.002 |
| 397 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.643 |
-0.058 |
| 398 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
-0.643 |
-0.344 |
| 399 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.642 |
-0.286 |
| 400 |
245399_at |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
-0.636 |
0.205 |
| 401 |
258235_at |
AT3G27620
|
AOX1C, alternative oxidase 1C |
-0.634 |
0.021 |
| 402 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.630 |
0.023 |
| 403 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.629 |
-0.110 |
| 404 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.628 |
0.058 |
| 405 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.626 |
-0.025 |
| 406 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.624 |
0.534 |
| 407 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
-0.623 |
-0.035 |
| 408 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.620 |
0.877 |
| 409 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
-0.618 |
0.287 |
| 410 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.617 |
0.312 |
| 411 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.611 |
0.036 |
| 412 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
-0.611 |
0.206 |
| 413 |
266009_at |
AT2G37420
|
[ATP binding microtubule motor family protein] |
-0.610 |
0.019 |
| 414 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.609 |
0.387 |
| 415 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.609 |
-0.198 |
| 416 |
249102_at |
AT5G43590
|
[Acyl transferase/acyl hydrolase/lysophospholipase superfamily protein] |
-0.607 |
-0.008 |
| 417 |
245440_at |
AT4G16680
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.603 |
-0.258 |
| 418 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.602 |
0.465 |
| 419 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.598 |
0.258 |
| 420 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
-0.597 |
0.193 |
| 421 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
-0.596 |
0.059 |
| 422 |
259878_at |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
-0.596 |
0.186 |
| 423 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.595 |
-0.260 |
| 424 |
251323_at |
AT3G61580
|
AtSLD1, SLD1, sphingoid LCB desaturase 1 |
-0.595 |
0.073 |
| 425 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.594 |
0.377 |
| 426 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.592 |
-0.023 |
| 427 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.591 |
-0.146 |
| 428 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.590 |
0.014 |
| 429 |
265927_at |
AT2G18590
|
[Major facilitator superfamily protein] |
-0.590 |
-0.066 |
| 430 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.589 |
1.304 |
| 431 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
-0.588 |
-0.116 |
| 432 |
257718_at |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
-0.584 |
0.191 |
| 433 |
258209_at |
AT3G14060
|
unknown |
-0.581 |
-0.081 |
| 434 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.574 |
-0.040 |
| 435 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
-0.570 |
0.217 |
| 436 |
254326_at |
AT4G22610
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.570 |
-0.088 |
| 437 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.568 |
-0.392 |
| 438 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.563 |
0.032 |
| 439 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.562 |
0.571 |
| 440 |
262399_at |
AT1G49500
|
unknown |
-0.562 |
-0.008 |
| 441 |
254193_at |
AT4G23870
|
unknown |
-0.561 |
0.135 |
| 442 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.559 |
0.135 |
| 443 |
258424_at |
AT3G16750
|
unknown |
-0.555 |
-0.039 |
| 444 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.555 |
-0.009 |
| 445 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.554 |
-0.121 |
| 446 |
265978_at |
AT2G11140
|
[copia-like retrotransposon family, has a 1.0e-71 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.550 |
-0.075 |
| 447 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.549 |
0.437 |
| 448 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.549 |
-0.293 |
| 449 |
247167_at |
AT5G65850
|
[F-box and associated interaction domains-containing protein] |
-0.549 |
0.158 |
| 450 |
248432_at |
AT5G51390
|
unknown |
-0.549 |
-0.031 |
| 451 |
259327_at |
AT3G16460
|
JAL34, jacalin-related lectin 34 |
-0.549 |
-0.086 |
| 452 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
-0.548 |
0.066 |
| 453 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.546 |
-0.103 |
| 454 |
263867_at |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
-0.545 |
0.001 |
| 455 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.544 |
0.543 |
| 456 |
265395_at |
AT2G20850
|
SRF1, STRUBBELIG-receptor family 1 |
-0.544 |
0.076 |
| 457 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.544 |
0.411 |
| 458 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.542 |
0.219 |
| 459 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.540 |
0.713 |
| 460 |
257974_at |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
-0.539 |
-0.076 |
| 461 |
256382_at |
AT1G66860
|
[Class I glutamine amidotransferase-like superfamily protein] |
-0.539 |
0.092 |
| 462 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.538 |
-0.123 |
| 463 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.534 |
0.044 |
| 464 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.534 |
0.325 |
| 465 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.533 |
0.145 |
| 466 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.531 |
-0.029 |
| 467 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
-0.528 |
0.140 |
| 468 |
258727_at |
AT3G11930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.527 |
0.021 |
| 469 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.525 |
-0.189 |
| 470 |
261145_at |
AT1G19730
|
ATH4, thioredoxin H-type 4, ATTRX4 |
-0.525 |
0.257 |
| 471 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.523 |
0.271 |
| 472 |
247210_at |
AT5G65020
|
ANNAT2, annexin 2, AtANN2 |
-0.522 |
0.405 |
| 473 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.521 |
-0.062 |
| 474 |
247328_at |
AT5G64130
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.521 |
0.148 |
| 475 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
-0.520 |
0.037 |
| 476 |
259288_at |
AT3G11500
|
[Small nuclear ribonucleoprotein family protein] |
-0.519 |
0.120 |
| 477 |
251012_at |
AT5G02580
|
unknown |
-0.516 |
-0.005 |
| 478 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.516 |
-0.045 |
| 479 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.515 |
0.288 |
| 480 |
247124_at |
AT5G66060
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.515 |
0.029 |
| 481 |
260841_at |
AT1G29195
|
unknown |
-0.512 |
0.090 |
| 482 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.511 |
0.218 |
| 483 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.510 |
0.471 |
| 484 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.510 |
0.253 |
| 485 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
-0.509 |
0.093 |
| 486 |
247344_at |
AT5G63750
|
ARI13, ARIADNE 13, ATARI13, ARABIDOPSIS ARIADNE 13 |
-0.509 |
-0.200 |
| 487 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.509 |
0.160 |
| 488 |
257450_at |
AT1G10530
|
unknown |
-0.508 |
-0.103 |
| 489 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
-0.508 |
-0.105 |
| 490 |
246699_at |
AT5G27990
|
[Pre-rRNA-processing protein TSR2, conserved region] |
-0.508 |
-0.179 |
| 491 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.508 |
0.025 |
| 492 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.507 |
0.186 |
| 493 |
249750_at |
AT5G24570
|
unknown |
-0.506 |
0.179 |
| 494 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.506 |
0.067 |
| 495 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.505 |
0.037 |
| 496 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
-0.503 |
0.002 |
| 497 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.502 |
-0.335 |
| 498 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.501 |
0.542 |
| 499 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.501 |
-0.004 |
| 500 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.500 |
0.627 |
| 501 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.499 |
-0.370 |
| 502 |
253155_at |
AT4G35720
|
unknown |
-0.498 |
0.263 |
| 503 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
-0.497 |
0.238 |
| 504 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.493 |
-0.332 |
| 505 |
246997_at |
AT5G67390
|
unknown |
-0.491 |
0.080 |
| 506 |
267428_at |
AT2G34840
|
[Coatomer epsilon subunit] |
-0.490 |
0.005 |
| 507 |
258953_at |
AT3G01430
|
unknown |
-0.490 |
-0.114 |
| 508 |
257798_at |
AT3G15950
|
NAI2 |
-0.489 |
-0.279 |
| 509 |
245127_at |
AT2G47600
|
ATMHX, magnesium/proton exchanger, ATMHX1, MHX, magnesium/proton exchanger, MHX1, MAGNESIUM/PROTON EXCHANGER 1 |
-0.488 |
0.089 |
| 510 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.484 |
0.124 |
| 511 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-0.483 |
0.039 |
| 512 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.482 |
0.635 |
| 513 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.481 |
0.169 |
| 514 |
260012_at |
AT1G67865
|
unknown |
-0.480 |
-0.145 |
| 515 |
266297_at |
AT2G29570
|
ATPCNA2, A. THALIANA PROLIFERATING CELL NUCLEAR ANTIGEN 2, PCNA2, proliferating cell nuclear antigen 2 |
-0.480 |
0.312 |
| 516 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.479 |
0.682 |
| 517 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.478 |
-0.074 |
| 518 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.477 |
0.012 |
| 519 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
-0.477 |
-0.153 |
| 520 |
251834_at |
AT3G55170
|
[Ribosomal L29 family protein ] |
-0.475 |
0.218 |
| 521 |
245301_at |
AT4G17190
|
FPS2, farnesyl diphosphate synthase 2 |
-0.474 |
0.402 |
| 522 |
254828_at |
AT4G12550
|
AIR1, Auxin-Induced in Root cultures 1 |
-0.474 |
-0.181 |
| 523 |
257757_at |
AT3G18660
|
GUX1, glucuronic acid substitution of xylan 1, PGSIP1, plant glycogenin-like starch initiation protein 1 |
-0.473 |
-0.561 |
| 524 |
252189_at |
AT3G50070
|
CYCD3;3, CYCLIN D3;3 |
-0.473 |
0.074 |
| 525 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.473 |
0.841 |
| 526 |
255260_at |
AT4G05040
|
[ankyrin repeat family protein] |
-0.472 |
0.009 |
| 527 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.471 |
-0.029 |
| 528 |
259637_at |
AT1G52260
|
ATPDI3, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 3, ATPDIL1-5, PDI-like 1-5, PDI3, PROTEIN DISULFIDE ISOMERASE 3, PDIL1-5, PDI-like 1-5 |
-0.471 |
-0.176 |
| 529 |
250441_at |
AT5G10540
|
TOP2, thimet metalloendopeptidase 2 |
-0.470 |
0.046 |
| 530 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.470 |
1.086 |
| 531 |
255285_at |
AT4G04630
|
unknown |
-0.470 |
-0.061 |
| 532 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.470 |
0.090 |
| 533 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.468 |
-0.440 |
| 534 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
-0.468 |
0.133 |
| 535 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.468 |
-0.114 |
| 536 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.467 |
0.023 |
| 537 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.467 |
-0.049 |
| 538 |
245261_at |
AT4G14385
|
unknown |
-0.464 |
-0.132 |
| 539 |
252211_at |
AT3G50220
|
IRX15, IRREGULAR XYLEM 15 |
-0.462 |
-0.417 |
| 540 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.462 |
0.060 |
| 541 |
265641_at |
AT2G27330
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.462 |
0.122 |
| 542 |
247706_at |
AT5G59480
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.458 |
0.247 |
| 543 |
245055_at |
AT2G26470
|
unknown |
-0.457 |
-0.062 |
| 544 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
-0.455 |
0.195 |
| 545 |
247151_at |
AT5G65640
|
bHLH093, beta HLH protein 93 |
-0.454 |
0.410 |
| 546 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
-0.453 |
0.238 |
| 547 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
-0.453 |
-0.038 |
| 548 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.452 |
0.936 |
| 549 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
-0.451 |
0.144 |
| 550 |
257141_at |
AT3G28900
|
[Ribosomal protein L34e superfamily protein] |
-0.451 |
0.353 |
| 551 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
-0.451 |
0.900 |
| 552 |
252852_at |
AT4G39900
|
unknown |
-0.451 |
-0.096 |
| 553 |
264954_at |
AT1G77060
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.450 |
-0.000 |
| 554 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
-0.450 |
-0.216 |
| 555 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
-0.448 |
0.193 |
| 556 |
245169_at |
AT2G33220
|
[GRIM-19 protein] |
-0.447 |
0.031 |
| 557 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.447 |
0.005 |
| 558 |
248977_at |
AT5G45020
|
[Glutathione S-transferase family protein] |
-0.446 |
-0.309 |
| 559 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
-0.446 |
-0.082 |
| 560 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
-0.445 |
0.113 |
| 561 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
-0.445 |
-0.046 |
| 562 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.445 |
-0.166 |
| 563 |
267627_at |
AT2G42270
|
[U5 small nuclear ribonucleoprotein helicase] |
-0.444 |
0.012 |
| 564 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
-0.443 |
-0.043 |
| 565 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.442 |
0.439 |
| 566 |
247111_at |
AT5G65880
|
unknown |
-0.442 |
0.037 |
| 567 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.441 |
0.093 |
| 568 |
256582_at |
AT3G28840
|
unknown |
-0.441 |
-0.047 |
| 569 |
245585_at |
AT4G14970
|
unknown |
-0.441 |
0.275 |
| 570 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
-0.441 |
0.540 |
| 571 |
260946_at |
AT1G06010
|
unknown |
-0.441 |
0.098 |
| 572 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.440 |
0.224 |
| 573 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.439 |
-0.006 |
| 574 |
257137_at |
AT3G28860
|
ABCB19, ATP-binding cassette B19, ATABCB19, Arabidopsis thaliana ATP-binding cassette B19, ATMDR1, ATMDR11, MULTIDRUG RESISTANCE PROTEIN 11, ATPGP19, MDR1, MDR11, PGP19, P-GLYCOPROTEIN 19 |
-0.437 |
0.231 |
| 575 |
261921_at |
AT1G65900
|
unknown |
-0.437 |
0.430 |
| 576 |
247988_at |
AT5G56910
|
[Proteinase inhibitor I25, cystatin, conserved region] |
-0.436 |
0.043 |
| 577 |
258641_at |
AT3G08030
|
unknown |
-0.436 |
0.029 |
| 578 |
261261_at |
AT1G26730
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.436 |
0.060 |
| 579 |
251369_at |
AT3G60480
|
unknown |
-0.436 |
-0.093 |
| 580 |
250470_at |
AT5G10160
|
[Thioesterase superfamily protein] |
-0.435 |
0.131 |
| 581 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.433 |
0.058 |
| 582 |
255038_at |
AT4G09510
|
A/N-InvI, alkaline/neutral invertase I, CINV2, cytosolic invertase 2 |
-0.432 |
-0.060 |
| 583 |
255763_at |
AT1G16730
|
unknown |
-0.432 |
0.209 |
| 584 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
-0.431 |
0.164 |
| 585 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
-0.431 |
0.095 |
| 586 |
252147_at |
AT3G51270
|
[protein serine/threonine kinases] |
-0.430 |
0.390 |
| 587 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.429 |
-0.016 |
| 588 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.429 |
0.067 |
| 589 |
262073_at |
AT1G59640
|
BPE, BIG PETAL, BPEp, BIG PETAL P, BPEub, BIG PETAL UB, ZCW32 |
-0.429 |
-0.134 |
| 590 |
266808_at |
AT2G29995
|
unknown |
-0.428 |
-0.211 |
| 591 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.428 |
-0.181 |
| 592 |
245555_at |
AT4G15390
|
[HXXXD-type acyl-transferase family protein] |
-0.427 |
-0.019 |
| 593 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.427 |
-0.062 |
| 594 |
255527_at |
AT4G02360
|
unknown |
-0.426 |
0.143 |
| 595 |
267054_at |
AT2G38370
|
unknown |
-0.425 |
0.334 |
| 596 |
260708_at |
AT1G32310
|
SAMBA, SAMBA |
-0.425 |
0.304 |
| 597 |
252880_at |
AT4G39730
|
PLAT1, PLAT domain protein 1 |
-0.425 |
0.320 |
| 598 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
-0.425 |
-0.302 |
| 599 |
264162_at |
AT1G65290
|
mtACP2, mitochondrial acyl carrier protein 2 |
-0.425 |
0.208 |
| 600 |
260441_at |
AT1G68260
|
ALT3, acyl-lipid thioesterase 3 |
-0.422 |
0.090 |