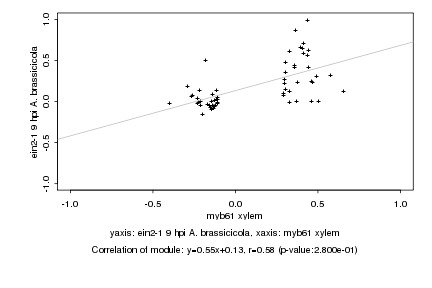
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
myb61 xylem |
Log2 signal ratio
ein2-1 9 hpi A. brassicicola |
| 1 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.651 |
0.123 |
| 2 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.575 |
0.317 |
| 3 |
253322_at |
AT4G33980
|
unknown |
0.498 |
0.005 |
| 4 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.489 |
0.316 |
| 5 |
253859_at |
AT4G27657
|
unknown |
0.463 |
0.237 |
| 6 |
257565_at |
AT3G28620
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
0.459 |
0.011 |
| 7 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.458 |
0.253 |
| 8 |
262211_at |
AT1G74930
|
ORA47 |
0.441 |
0.426 |
| 9 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.439 |
0.622 |
| 10 |
256442_at |
AT3G10930
|
unknown |
0.432 |
0.998 |
| 11 |
253643_at |
AT4G29780
|
unknown |
0.432 |
0.566 |
| 12 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.411 |
0.588 |
| 13 |
247047_at |
AT5G66650
|
unknown |
0.411 |
0.711 |
| 14 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.404 |
0.650 |
| 15 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.389 |
0.661 |
| 16 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.371 |
0.233 |
| 17 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.364 |
0.007 |
| 18 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.363 |
0.874 |
| 19 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.357 |
0.426 |
| 20 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.357 |
0.443 |
| 21 |
260744_at |
AT1G15010
|
unknown |
0.327 |
0.130 |
| 22 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.326 |
0.615 |
| 23 |
260227_at |
AT1G74450
|
unknown |
0.323 |
-0.010 |
| 24 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.302 |
0.148 |
| 25 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.301 |
0.480 |
| 26 |
265184_at |
AT1G23710
|
unknown |
0.298 |
0.363 |
| 27 |
253832_at |
AT4G27654
|
unknown |
0.295 |
0.270 |
| 28 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.292 |
0.220 |
| 29 |
261405_at |
AT1G18740
|
unknown |
0.290 |
0.074 |
| 30 |
257925_at |
AT3G23170
|
unknown |
0.289 |
0.108 |
| 31 |
263292_at |
AT2G10900
|
[expressed protein] |
-0.405 |
-0.016 |
| 32 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.291 |
0.190 |
| 33 |
262719_at |
AT1G43590
|
unknown |
-0.270 |
0.069 |
| 34 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.263 |
0.080 |
| 35 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
-0.235 |
-0.016 |
| 36 |
247051_at |
AT5G66670
|
unknown |
-0.234 |
0.040 |
| 37 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.225 |
0.000 |
| 38 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.220 |
0.139 |
| 39 |
248582_at |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.216 |
-0.046 |
| 40 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-0.213 |
0.005 |
| 41 |
256518_at |
AT1G66080
|
unknown |
-0.202 |
-0.152 |
| 42 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.187 |
0.505 |
| 43 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
-0.173 |
-0.025 |
| 44 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-0.161 |
-0.040 |
| 45 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.156 |
-0.070 |
| 46 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
-0.149 |
0.008 |
| 47 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.147 |
-0.095 |
| 48 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
-0.145 |
-0.040 |
| 49 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
-0.140 |
0.093 |
| 50 |
247522_at |
AT5G61340
|
unknown |
-0.136 |
-0.081 |
| 51 |
253532_at |
AT4G31570
|
unknown |
-0.134 |
-0.059 |
| 52 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.131 |
-0.051 |
| 53 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
-0.127 |
0.014 |
| 54 |
257224_at |
AT3G27870
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.123 |
-0.044 |
| 55 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
-0.119 |
0.137 |
| 56 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.118 |
0.034 |
| 57 |
256778_at |
AT3G13782
|
NAP1;4, nucleosome assembly protein1;4, NFA04, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 04, NFA4, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 4 |
-0.114 |
0.035 |
| 58 |
266717_at |
AT2G46735
|
unknown |
-0.112 |
-0.020 |
| 59 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.111 |
-0.004 |
| 60 |
259766_at |
AT1G64360
|
unknown |
-0.109 |
0.052 |