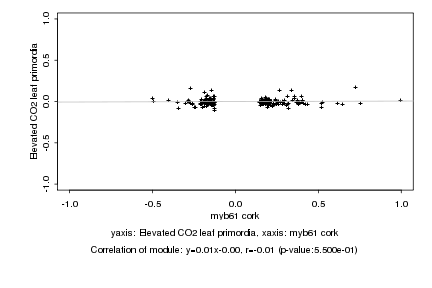
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
myb61 cork |
Log2 signal ratio
Elevated CO2 leaf primordia |
| 1 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.993 |
0.020 |
| 2 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.754 |
-0.014 |
| 3 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.721 |
0.181 |
| 4 |
256009_at |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
0.642 |
-0.032 |
| 5 |
262211_at |
AT1G74930
|
ORA47 |
0.615 |
-0.022 |
| 6 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.520 |
-0.011 |
| 7 |
253643_at |
AT4G29780
|
unknown |
0.517 |
-0.023 |
| 8 |
253859_at |
AT4G27657
|
unknown |
0.514 |
-0.070 |
| 9 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.432 |
-0.036 |
| 10 |
247789_at |
AT5G58680
|
[ARM repeat superfamily protein] |
0.421 |
-0.029 |
| 11 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.407 |
-0.022 |
| 12 |
266316_at |
AT2G27080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.404 |
0.018 |
| 13 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
0.399 |
0.023 |
| 14 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.397 |
0.070 |
| 15 |
256442_at |
AT3G10930
|
unknown |
0.393 |
-0.003 |
| 16 |
251192_at |
AT3G62720
|
ATXT1, XT1, xylosyltransferase 1, XXT1, XYG XYLOSYLTRANSFERASE 1 |
0.385 |
-0.009 |
| 17 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.381 |
-0.012 |
| 18 |
260227_at |
AT1G74450
|
unknown |
0.378 |
-0.030 |
| 19 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.376 |
-0.004 |
| 20 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.374 |
0.018 |
| 21 |
250098_at |
AT5G17350
|
unknown |
0.373 |
-0.007 |
| 22 |
260135_at |
AT1G66400
|
CML23, calmodulin like 23 |
0.370 |
-0.002 |
| 23 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.363 |
0.000 |
| 24 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
0.355 |
0.038 |
| 25 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.354 |
0.061 |
| 26 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.339 |
0.013 |
| 27 |
261684_at |
AT1G47400
|
unknown |
0.337 |
0.139 |
| 28 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.319 |
-0.073 |
| 29 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.315 |
-0.022 |
| 30 |
261405_at |
AT1G18740
|
unknown |
0.312 |
-0.028 |
| 31 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.309 |
0.069 |
| 32 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.302 |
-0.046 |
| 33 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.295 |
-0.015 |
| 34 |
259729_at |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
0.291 |
0.021 |
| 35 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.280 |
-0.036 |
| 36 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
0.279 |
0.004 |
| 37 |
265732_at |
AT2G01300
|
unknown |
0.278 |
-0.005 |
| 38 |
267515_at |
AT2G45680
|
TCP9, TCP domain protein 9 |
0.268 |
-0.000 |
| 39 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.260 |
0.145 |
| 40 |
253832_at |
AT4G27654
|
unknown |
0.259 |
-0.029 |
| 41 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.258 |
-0.011 |
| 42 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.255 |
0.007 |
| 43 |
250676_at |
AT5G06320
|
NHL3, NDR1/HIN1-like 3 |
0.249 |
-0.003 |
| 44 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
0.247 |
0.012 |
| 45 |
247708_at |
AT5G59550
|
unknown |
0.245 |
-0.016 |
| 46 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.243 |
-0.026 |
| 47 |
259979_at |
AT1G76600
|
unknown |
0.239 |
0.032 |
| 48 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.236 |
0.021 |
| 49 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.226 |
-0.045 |
| 50 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.226 |
-0.027 |
| 51 |
255733_at |
AT1G25400
|
unknown |
0.225 |
-0.021 |
| 52 |
252474_at |
AT3G46620
|
unknown |
0.224 |
-0.019 |
| 53 |
263931_at |
AT2G36220
|
unknown |
0.218 |
-0.059 |
| 54 |
259105_at |
AT3G05500
|
[Rubber elongation factor protein (REF)] |
0.217 |
0.010 |
| 55 |
246584_at |
AT5G14730
|
unknown |
0.212 |
0.001 |
| 56 |
251640_at |
AT3G57450
|
unknown |
0.207 |
0.006 |
| 57 |
253830_at |
AT4G27652
|
unknown |
0.206 |
-0.041 |
| 58 |
266037_at |
AT2G05940
|
RIPK, RPM1-induced protein kinase |
0.205 |
0.009 |
| 59 |
260367_at |
AT1G69760
|
unknown |
0.201 |
0.026 |
| 60 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.200 |
-0.017 |
| 61 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.200 |
0.003 |
| 62 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.199 |
-0.041 |
| 63 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.198 |
0.032 |
| 64 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.196 |
0.029 |
| 65 |
251366_at |
AT3G61340
|
[F-box and associated interaction domains-containing protein] |
0.196 |
-0.033 |
| 66 |
262225_at |
AT1G53840
|
ATPME1, pectin methylesterase 1, PME1, pectin methylesterase 1 |
0.195 |
-0.021 |
| 67 |
260686_at |
AT1G17620
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.194 |
-0.017 |
| 68 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.192 |
-0.002 |
| 69 |
260075_at |
AT1G73700
|
[MATE efflux family protein] |
0.189 |
-0.063 |
| 70 |
261115_at |
AT1G75360
|
unknown |
0.187 |
0.015 |
| 71 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
0.186 |
0.047 |
| 72 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.184 |
0.026 |
| 73 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.181 |
0.056 |
| 74 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.180 |
-0.015 |
| 75 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.179 |
-0.028 |
| 76 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.176 |
0.018 |
| 77 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
0.175 |
0.022 |
| 78 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
0.174 |
0.000 |
| 79 |
250024_at |
AT5G18270
|
ANAC087, Arabidopsis NAC domain containing protein 87 |
0.170 |
-0.004 |
| 80 |
246388_at |
AT1G77405
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.170 |
-0.015 |
| 81 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.165 |
-0.029 |
| 82 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
0.163 |
-0.023 |
| 83 |
260211_at |
AT1G74440
|
unknown |
0.162 |
0.016 |
| 84 |
261193_at |
AT1G32920
|
unknown |
0.160 |
0.002 |
| 85 |
253925_at |
AT4G26690
|
GDPDL3, Glycerophosphodiester phosphodiesterase (GDPD) like 3, GPDL2, GLYCEROPHOSPHODIESTERASE-LIKE 2, MRH5, MUTANT ROOT HAIR 5, SHV3, SHAVEN 3 |
0.158 |
-0.024 |
| 86 |
256361_at |
AT1G66520
|
pde194, pigment defective 194 |
0.158 |
0.036 |
| 87 |
263613_at |
AT2G25250
|
unknown |
0.156 |
-0.029 |
| 88 |
250018_at |
AT5G18150
|
[Methyltransferase-related protein] |
0.156 |
-0.014 |
| 89 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.155 |
0.038 |
| 90 |
253638_at |
AT4G30470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.151 |
-0.044 |
| 91 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.148 |
0.007 |
| 92 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
0.144 |
-0.003 |
| 93 |
254845_at |
AT4G11820
|
FKP1, FLAKY POLLEN 1, HMGS, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, MVA1 |
0.144 |
0.007 |
| 94 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.143 |
0.003 |
| 95 |
263292_at |
AT2G10900
|
[expressed protein] |
-0.505 |
0.047 |
| 96 |
257323_at |
ATMG01200
|
ORF294 |
-0.500 |
0.006 |
| 97 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.404 |
0.015 |
| 98 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.354 |
-0.002 |
| 99 |
259766_at |
AT1G64360
|
unknown |
-0.349 |
-0.078 |
| 100 |
262719_at |
AT1G43590
|
unknown |
-0.303 |
-0.016 |
| 101 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.292 |
0.001 |
| 102 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.289 |
0.023 |
| 103 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.285 |
0.001 |
| 104 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.278 |
0.005 |
| 105 |
260149_at |
AT1G52810
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.276 |
-0.012 |
| 106 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.275 |
0.158 |
| 107 |
262656_at |
AT1G14200
|
[RING/U-box superfamily protein] |
-0.260 |
-0.020 |
| 108 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
-0.259 |
-0.034 |
| 109 |
261221_at |
AT1G19960
|
unknown |
-0.250 |
-0.062 |
| 110 |
249010_at |
AT5G44580
|
unknown |
-0.243 |
-0.065 |
| 111 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.216 |
0.004 |
| 112 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.216 |
-0.015 |
| 113 |
259235_at |
AT3G11600
|
unknown |
-0.212 |
-0.030 |
| 114 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
-0.209 |
-0.029 |
| 115 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.207 |
-0.017 |
| 116 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.206 |
0.034 |
| 117 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
-0.206 |
-0.014 |
| 118 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
-0.203 |
-0.005 |
| 119 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.199 |
-0.071 |
| 120 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-0.193 |
-0.010 |
| 121 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.193 |
0.009 |
| 122 |
260005_at |
AT1G67920
|
unknown |
-0.192 |
0.115 |
| 123 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.192 |
-0.019 |
| 124 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
-0.189 |
-0.049 |
| 125 |
245004_at |
ATCG00300
|
YCF9 |
-0.188 |
-0.011 |
| 126 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
-0.188 |
0.012 |
| 127 |
257153_at |
AT3G27220
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.184 |
0.027 |
| 128 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
-0.184 |
-0.018 |
| 129 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-0.181 |
-0.017 |
| 130 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.180 |
-0.053 |
| 131 |
266717_at |
AT2G46735
|
unknown |
-0.179 |
-0.012 |
| 132 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.177 |
0.043 |
| 133 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.176 |
0.062 |
| 134 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.175 |
0.034 |
| 135 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.174 |
-0.017 |
| 136 |
260264_at |
AT1G68500
|
unknown |
-0.173 |
-0.007 |
| 137 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.173 |
-0.024 |
| 138 |
261567_at |
AT1G33055
|
unknown |
-0.171 |
0.085 |
| 139 |
257946_at |
AT3G21710
|
unknown |
-0.171 |
-0.015 |
| 140 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
-0.169 |
-0.019 |
| 141 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
-0.169 |
-0.008 |
| 142 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.166 |
-0.015 |
| 143 |
267279_at |
AT2G19460
|
unknown |
-0.166 |
-0.024 |
| 144 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.166 |
0.012 |
| 145 |
245353_at |
AT4G16000
|
unknown |
-0.161 |
0.047 |
| 146 |
260332_at |
AT1G70470
|
unknown |
-0.160 |
0.010 |
| 147 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.160 |
0.024 |
| 148 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.159 |
0.036 |
| 149 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.155 |
-0.034 |
| 150 |
246946_at |
AT5G25070
|
unknown |
-0.154 |
-0.012 |
| 151 |
253567_at |
AT4G31230
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.153 |
0.001 |
| 152 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
-0.153 |
0.020 |
| 153 |
256824_at |
AT3G22121
|
other RNA |
-0.153 |
-0.021 |
| 154 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.153 |
0.042 |
| 155 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.153 |
0.001 |
| 156 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.151 |
0.033 |
| 157 |
261073_at |
AT1G07300
|
[josephin protein-related] |
-0.150 |
-0.020 |
| 158 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.149 |
-0.026 |
| 159 |
263603_at |
AT2G16340
|
unknown |
-0.148 |
-0.039 |
| 160 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.148 |
0.143 |
| 161 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.147 |
0.006 |
| 162 |
245953_at |
AT5G28520
|
[Mannose-binding lectin superfamily protein] |
-0.147 |
0.005 |
| 163 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.147 |
-0.033 |
| 164 |
247051_at |
AT5G66670
|
unknown |
-0.145 |
-0.014 |
| 165 |
265769_at |
AT2G48090
|
unknown |
-0.145 |
0.033 |
| 166 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.143 |
-0.018 |
| 167 |
247072_at |
AT5G66490
|
unknown |
-0.142 |
-0.046 |
| 168 |
245330_at |
AT4G14930
|
[Survival protein SurE-like phosphatase/nucleotidase] |
-0.141 |
-0.004 |
| 169 |
262602_at |
AT1G15270
|
[Translation machinery associated TMA7] |
-0.139 |
-0.020 |
| 170 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.138 |
0.063 |
| 171 |
262151_at |
AT1G52510
|
[alpha/beta-Hydrolases superfamily protein] |
-0.136 |
-0.013 |
| 172 |
258262_at |
AT3G15770
|
unknown |
-0.135 |
-0.039 |
| 173 |
252198_x_at |
AT3G50250
|
unknown |
-0.135 |
-0.017 |
| 174 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-0.135 |
0.048 |
| 175 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.133 |
0.046 |
| 176 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.133 |
0.025 |
| 177 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
-0.133 |
-0.009 |
| 178 |
248969_at |
AT5G45310
|
unknown |
-0.133 |
-0.048 |
| 179 |
264621_at |
AT2G17700
|
STY8, serine/threonine/tyrosine kinase 8 |
-0.133 |
0.006 |
| 180 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.133 |
0.002 |
| 181 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
-0.132 |
-0.097 |
| 182 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
-0.131 |
-0.004 |
| 183 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.131 |
0.069 |
| 184 |
249995_at |
AT5G18610
|
[Protein kinase superfamily protein] |
-0.131 |
0.006 |
| 185 |
256569_at |
AT3G19550
|
unknown |
-0.130 |
-0.024 |
| 186 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.129 |
-0.076 |
| 187 |
253432_at |
AT4G32450
|
MEF8S, MEF8 similar |
-0.129 |
-0.011 |