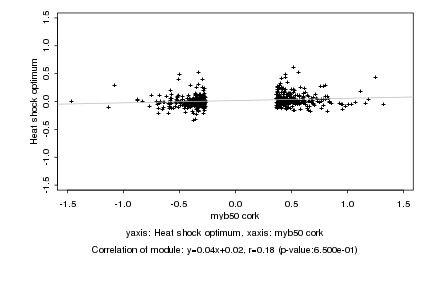
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
myb50 cork |
Log2 signal ratio
Heat shock optimum |
| 1 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
1.320 |
-0.045 |
| 2 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
1.250 |
0.443 |
| 3 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
1.183 |
0.040 |
| 4 |
250472_at |
AT5G10210
|
unknown |
1.157 |
-0.029 |
| 5 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
1.117 |
0.191 |
| 6 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
1.072 |
-0.008 |
| 7 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
1.034 |
-0.039 |
| 8 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
1.006 |
-0.048 |
| 9 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.983 |
-0.087 |
| 10 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
0.951 |
-0.137 |
| 11 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.948 |
-0.053 |
| 12 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.942 |
-0.039 |
| 13 |
262399_at |
AT1G49500
|
unknown |
0.924 |
-0.022 |
| 14 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
0.857 |
-0.019 |
| 15 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
0.837 |
-0.004 |
| 16 |
259975_at |
AT1G76470
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.836 |
0.006 |
| 17 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.829 |
0.071 |
| 18 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.819 |
0.094 |
| 19 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.818 |
-0.178 |
| 20 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.803 |
0.297 |
| 21 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
0.800 |
0.096 |
| 22 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.796 |
0.038 |
| 23 |
252882_at |
AT4G39675
|
unknown |
0.785 |
-0.064 |
| 24 |
265117_at |
AT1G62500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.784 |
0.284 |
| 25 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.779 |
0.054 |
| 26 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.763 |
0.047 |
| 27 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.761 |
-0.124 |
| 28 |
247037_at |
AT5G67070
|
RALFL34, ralf-like 34 |
0.760 |
0.004 |
| 29 |
256125_at |
AT1G18250
|
ATLP-1 |
0.752 |
0.276 |
| 30 |
247024_at |
AT5G66985
|
unknown |
0.742 |
-0.008 |
| 31 |
266549_at |
AT2G35150
|
EXL7, EXORDIUM LIKE 7 |
0.733 |
0.017 |
| 32 |
257761_at |
AT3G23090
|
WDL3 |
0.727 |
0.054 |
| 33 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
0.717 |
0.055 |
| 34 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
0.710 |
0.140 |
| 35 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.698 |
-0.066 |
| 36 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.696 |
-0.021 |
| 37 |
261363_at |
AT1G41830
|
SKS6, SKU5 SIMILAR 6, SKS6, SKU5-similar 6 |
0.695 |
-0.041 |
| 38 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.690 |
0.085 |
| 39 |
265204_at |
AT2G36650
|
unknown |
0.681 |
0.025 |
| 40 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
0.679 |
0.058 |
| 41 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.675 |
0.014 |
| 42 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
0.672 |
0.068 |
| 43 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
0.671 |
-0.015 |
| 44 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.663 |
-0.178 |
| 45 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.653 |
-0.082 |
| 46 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
0.653 |
0.006 |
| 47 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.649 |
0.094 |
| 48 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.645 |
0.009 |
| 49 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
0.644 |
-0.159 |
| 50 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.642 |
-0.111 |
| 51 |
261930_at |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.641 |
-0.073 |
| 52 |
261921_at |
AT1G65900
|
unknown |
0.641 |
0.115 |
| 53 |
245226_at |
AT3G29970
|
[B12D protein] |
0.623 |
-0.051 |
| 54 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.621 |
0.168 |
| 55 |
253317_at |
AT4G33960
|
unknown |
0.617 |
-0.013 |
| 56 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.616 |
0.248 |
| 57 |
255876_at |
AT2G40480
|
unknown |
0.613 |
0.031 |
| 58 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.612 |
-0.002 |
| 59 |
254361_at |
AT4G22212
|
[Arabidopsis defensin-like protein] |
0.607 |
0.075 |
| 60 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.606 |
0.026 |
| 61 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.603 |
0.106 |
| 62 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.602 |
0.161 |
| 63 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.600 |
0.019 |
| 64 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.600 |
-0.023 |
| 65 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.596 |
0.000 |
| 66 |
251420_at |
AT3G60490
|
[Integrase-type DNA-binding superfamily protein] |
0.594 |
0.060 |
| 67 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
0.586 |
-0.013 |
| 68 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
0.579 |
-0.143 |
| 69 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.576 |
0.258 |
| 70 |
264611_at |
AT1G04680
|
[Pectin lyase-like superfamily protein] |
0.573 |
-0.024 |
| 71 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.570 |
-0.049 |
| 72 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
0.569 |
0.094 |
| 73 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
0.566 |
0.024 |
| 74 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
0.566 |
0.034 |
| 75 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
0.566 |
0.110 |
| 76 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.564 |
-0.034 |
| 77 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.562 |
0.528 |
| 78 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.559 |
-0.002 |
| 79 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
0.555 |
-0.010 |
| 80 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.553 |
0.118 |
| 81 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.553 |
-0.052 |
| 82 |
258641_at |
AT3G08030
|
unknown |
0.552 |
0.033 |
| 83 |
248763_at |
AT5G47550
|
[Cystatin/monellin superfamily protein] |
0.550 |
-0.040 |
| 84 |
254815_at |
AT4G12420
|
SKU5 |
0.547 |
0.002 |
| 85 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.546 |
-0.007 |
| 86 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
0.546 |
0.111 |
| 87 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
0.538 |
0.008 |
| 88 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
0.535 |
-0.035 |
| 89 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.532 |
-0.048 |
| 90 |
261521_at |
AT1G71830
|
ATSERK1, SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1, SERK1, somatic embryogenesis receptor-like kinase 1 |
0.531 |
0.047 |
| 91 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
0.531 |
-0.010 |
| 92 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
0.530 |
0.036 |
| 93 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.528 |
0.112 |
| 94 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.527 |
-0.145 |
| 95 |
247245_at |
AT5G64720
|
EC1.5, EGG CELL 1.5 |
0.524 |
-0.034 |
| 96 |
255104_at |
AT4G08685
|
SAH7 |
0.523 |
0.024 |
| 97 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.523 |
0.025 |
| 98 |
261460_at |
AT1G07880
|
ATMPK13 |
0.521 |
0.070 |
| 99 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.520 |
0.200 |
| 100 |
257617_at |
AT3G26550
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.519 |
-0.031 |
| 101 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.519 |
0.032 |
| 102 |
261346_at |
AT1G79720
|
[Eukaryotic aspartyl protease family protein] |
0.518 |
-0.018 |
| 103 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
0.518 |
0.087 |
| 104 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.517 |
-0.008 |
| 105 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.516 |
0.220 |
| 106 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.516 |
0.175 |
| 107 |
267459_at |
AT2G33850
|
unknown |
0.514 |
0.616 |
| 108 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
0.512 |
-0.157 |
| 109 |
257815_at |
AT3G25130
|
unknown |
0.507 |
-0.005 |
| 110 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.506 |
0.026 |
| 111 |
252501_at |
AT3G46880
|
unknown |
0.505 |
0.001 |
| 112 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.504 |
0.072 |
| 113 |
261728_at |
AT1G76160
|
sks5, SKU5 similar 5 |
0.504 |
0.032 |
| 114 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.504 |
-0.084 |
| 115 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.502 |
0.040 |
| 116 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.501 |
0.053 |
| 117 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.500 |
0.040 |
| 118 |
263496_at |
AT2G42570
|
TBL39, TRICHOME BIREFRINGENCE-LIKE 39 |
0.499 |
0.088 |
| 119 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
0.497 |
0.218 |
| 120 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
0.496 |
0.068 |
| 121 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
0.493 |
0.079 |
| 122 |
248139_at |
AT5G54970
|
unknown |
0.492 |
0.056 |
| 123 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
0.492 |
-0.008 |
| 124 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.491 |
0.152 |
| 125 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
0.491 |
-0.024 |
| 126 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
0.490 |
-0.116 |
| 127 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
0.490 |
0.136 |
| 128 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.490 |
0.137 |
| 129 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.490 |
0.026 |
| 130 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.490 |
0.071 |
| 131 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.489 |
0.000 |
| 132 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
0.484 |
-0.016 |
| 133 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.482 |
0.094 |
| 134 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
0.481 |
-0.015 |
| 135 |
261533_at |
AT1G71690
|
unknown |
0.481 |
-0.005 |
| 136 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
0.480 |
-0.029 |
| 137 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
0.480 |
0.146 |
| 138 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.479 |
0.021 |
| 139 |
262980_at |
AT1G75680
|
AtGH9B7, glycosyl hydrolase 9B7, GH9B7, glycosyl hydrolase 9B7 |
0.479 |
-0.002 |
| 140 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
0.478 |
0.041 |
| 141 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
0.477 |
0.038 |
| 142 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
0.476 |
-0.017 |
| 143 |
266223_at |
AT2G28790
|
[Pathogenesis-related thaumatin superfamily protein] |
0.475 |
0.112 |
| 144 |
257918_at |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
0.473 |
0.014 |
| 145 |
251058_at |
AT5G01790
|
unknown |
0.472 |
-0.105 |
| 146 |
250337_at |
AT5G11790
|
NDL2, N-MYC downregulated-like 2 |
0.471 |
-0.052 |
| 147 |
263628_at |
AT2G04780
|
FLA7, FASCICLIN-like arabinoogalactan 7 |
0.471 |
-0.006 |
| 148 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
0.470 |
-0.033 |
| 149 |
246911_at |
AT5G25810
|
tny, TINY |
0.470 |
-0.010 |
| 150 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.469 |
0.111 |
| 151 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.468 |
0.156 |
| 152 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
0.466 |
-0.032 |
| 153 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.466 |
-0.035 |
| 154 |
245980_at |
AT5G13140
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.466 |
0.029 |
| 155 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
0.464 |
-0.093 |
| 156 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.462 |
0.006 |
| 157 |
258859_at |
AT3G02120
|
[hydroxyproline-rich glycoprotein family protein] |
0.462 |
0.354 |
| 158 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.461 |
0.102 |
| 159 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
0.459 |
-0.100 |
| 160 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.458 |
-0.032 |
| 161 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
0.458 |
0.128 |
| 162 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
0.457 |
0.031 |
| 163 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.456 |
0.041 |
| 164 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.456 |
-0.127 |
| 165 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
0.456 |
-0.005 |
| 166 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.455 |
-0.047 |
| 167 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
0.453 |
0.127 |
| 168 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
0.452 |
-0.038 |
| 169 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
0.450 |
0.060 |
| 170 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
0.449 |
-0.077 |
| 171 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
0.447 |
-0.019 |
| 172 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.445 |
0.057 |
| 173 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
0.444 |
-0.037 |
| 174 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
0.443 |
-0.049 |
| 175 |
260744_at |
AT1G15010
|
unknown |
0.443 |
0.501 |
| 176 |
255517_at |
AT4G02290
|
AtGH9B13, glycosyl hydrolase 9B13, GH9B13, glycosyl hydrolase 9B13 |
0.442 |
0.226 |
| 177 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.441 |
-0.117 |
| 178 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.441 |
0.026 |
| 179 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.441 |
0.433 |
| 180 |
256211_at |
AT1G50960
|
ATGA2OX7, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 7, GA2OX7, gibberellin 2-oxidase 7 |
0.438 |
-0.020 |
| 181 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
0.438 |
0.033 |
| 182 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.435 |
-0.045 |
| 183 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.434 |
0.186 |
| 184 |
260112_at |
AT1G63310
|
unknown |
0.434 |
0.034 |
| 185 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.434 |
0.009 |
| 186 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.433 |
-0.000 |
| 187 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.432 |
-0.069 |
| 188 |
255064_at |
AT4G08950
|
EXO, EXORDIUM |
0.431 |
0.177 |
| 189 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
0.431 |
-0.027 |
| 190 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
0.431 |
-0.036 |
| 191 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.430 |
0.068 |
| 192 |
249167_at |
AT5G42860
|
unknown |
0.430 |
0.080 |
| 193 |
261501_at |
AT1G28390
|
[Protein kinase superfamily protein] |
0.430 |
0.015 |
| 194 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.429 |
-0.044 |
| 195 |
251788_at |
AT3G55420
|
unknown |
0.428 |
0.053 |
| 196 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
0.427 |
-0.004 |
| 197 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
0.426 |
0.070 |
| 198 |
258480_at |
AT3G02640
|
unknown |
0.425 |
0.250 |
| 199 |
247850_at |
AT5G58150
|
[Leucine-rich repeat protein kinase family protein] |
0.425 |
-0.063 |
| 200 |
253010_at |
AT4G37750
|
ANT, AINTEGUMENTA, CKC, CKC1, COMPLEMENTING A PROTEIN KINASE C MUTANT 1, DRG, DRAGON |
0.425 |
0.132 |
| 201 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
0.423 |
0.050 |
| 202 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
0.422 |
0.076 |
| 203 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
0.421 |
0.089 |
| 204 |
248085_at |
AT5G55480
|
GDPDL4, Glycerophosphodiester phosphodiesterase (GDPD) like 4, GPDL1, glycerophosphodiesterase-like 1, SVL1, SHV3-like 1 |
0.421 |
0.010 |
| 205 |
259682_at |
AT1G63040
|
[pseudogene] |
0.420 |
-0.009 |
| 206 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
0.420 |
-0.039 |
| 207 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.419 |
-0.025 |
| 208 |
248154_at |
AT5G54400
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.418 |
0.041 |
| 209 |
265083_at |
AT1G03820
|
unknown |
0.417 |
0.061 |
| 210 |
252852_at |
AT4G39900
|
unknown |
0.417 |
0.057 |
| 211 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.416 |
0.235 |
| 212 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.415 |
-0.051 |
| 213 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.414 |
0.216 |
| 214 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
0.412 |
0.022 |
| 215 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.412 |
-0.021 |
| 216 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
0.411 |
0.076 |
| 217 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.411 |
-0.099 |
| 218 |
253286_at |
AT4G34260
|
AXY8, ALTERED XYLOGLUCAN 8, FUC95A |
0.410 |
0.027 |
| 219 |
262978_at |
AT1G75780
|
TUB1, tubulin beta-1 chain |
0.410 |
0.429 |
| 220 |
266227_at |
AT2G28870
|
unknown |
0.409 |
-0.122 |
| 221 |
251219_at |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
0.408 |
-0.037 |
| 222 |
259806_at |
AT1G47900
|
unknown |
0.408 |
0.005 |
| 223 |
259014_at |
AT3G07320
|
[O-Glycosyl hydrolases family 17 protein] |
0.408 |
0.085 |
| 224 |
260074_at |
AT1G73640
|
AtRABA6a, RAB GTPase homolog A6A, RABA6a, RAB GTPase homolog A6A |
0.407 |
0.021 |
| 225 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
0.407 |
0.071 |
| 226 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
0.407 |
-0.015 |
| 227 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.406 |
0.266 |
| 228 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.406 |
0.020 |
| 229 |
267355_at |
AT2G39900
|
WLIM2a, WLIM2a |
0.405 |
0.170 |
| 230 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
0.404 |
0.044 |
| 231 |
266325_at |
AT2G46630
|
unknown |
0.404 |
-0.043 |
| 232 |
258414_at |
AT3G17380
|
[TRAF-like family protein] |
0.404 |
0.012 |
| 233 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.404 |
0.007 |
| 234 |
257517_at |
AT3G16330
|
unknown |
0.403 |
0.030 |
| 235 |
249911_at |
AT5G22740
|
ATCSLA02, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A02, ATCSLA2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE A2, CSLA02, cellulose synthase-like A02, CSLA2, CELLULOSE SYNTHASE-LIKE A 2 |
0.403 |
0.065 |
| 236 |
257952_at |
AT3G21770
|
[Peroxidase superfamily protein] |
0.400 |
0.087 |
| 237 |
247425_at |
AT5G62550
|
unknown |
0.400 |
0.341 |
| 238 |
262225_at |
AT1G53840
|
ATPME1, pectin methylesterase 1, PME1, pectin methylesterase 1 |
0.399 |
-0.002 |
| 239 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
0.399 |
0.026 |
| 240 |
254418_at |
AT4G21480
|
STP12, sugar transporter protein 12 |
0.397 |
0.074 |
| 241 |
266477_at |
AT2G31085
|
AtCLE6, CLE6, CLAVATA3/ESR-RELATED 6 |
0.396 |
-0.013 |
| 242 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.396 |
0.011 |
| 243 |
261492_at |
AT1G14290
|
SBH2, sphingoid base hydroxylase 2 |
0.395 |
-0.000 |
| 244 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
0.394 |
0.007 |
| 245 |
257297_at |
AT3G28040
|
[Leucine-rich receptor-like protein kinase family protein] |
0.392 |
-0.061 |
| 246 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
0.392 |
-0.065 |
| 247 |
255851_at |
AT1G67040
|
TRM22, TON1 Recruiting Motif 22 |
0.392 |
-0.015 |
| 248 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.391 |
0.237 |
| 249 |
254448_at |
AT4G20870
|
ATFAH2, ARABIDOPSIS FATTY ACID HYDROXYLASE 2, FAH2, fatty acid hydroxylase 2 |
0.391 |
-0.005 |
| 250 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
0.391 |
0.288 |
| 251 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
0.391 |
0.006 |
| 252 |
259664_at |
AT1G55330
|
AGP21, arabinogalactan protein 21, ATAGP21, ARABINOGALACTAN PROTEIN 21 |
0.391 |
0.021 |
| 253 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.390 |
0.091 |
| 254 |
266425_at |
AT2G41300
|
SSL1, strictosidine synthase-like 1 |
0.390 |
0.003 |
| 255 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.390 |
-0.061 |
| 256 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
0.388 |
0.140 |
| 257 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
0.386 |
0.055 |
| 258 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.385 |
0.248 |
| 259 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
0.385 |
-0.027 |
| 260 |
249941_at |
AT5G22270
|
unknown |
0.385 |
-0.074 |
| 261 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
0.385 |
0.003 |
| 262 |
265957_at |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
0.384 |
0.095 |
| 263 |
251852_at |
AT3G54750
|
unknown |
0.383 |
0.059 |
| 264 |
262210_at |
AT1G74690
|
IQD31, IQ-domain 31 |
0.383 |
0.008 |
| 265 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.383 |
-0.061 |
| 266 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.382 |
-0.056 |
| 267 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
0.381 |
0.091 |
| 268 |
254372_at |
AT4G21620
|
[glycine-rich protein] |
0.381 |
0.062 |
| 269 |
257697_at |
AT3G12700
|
NANA, NANA |
0.381 |
0.054 |
| 270 |
263219_at |
AT1G30600
|
[Subtilase family protein] |
0.380 |
0.051 |
| 271 |
261223_at |
AT1G19950
|
HVA22H, HVA22-like protein H (ATHVA22H) |
0.378 |
-0.036 |
| 272 |
248861_at |
AT5G46700
|
TET1, TETRASPANIN 1, TRN2, TORNADO 2 |
0.378 |
0.057 |
| 273 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.378 |
0.222 |
| 274 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.377 |
0.075 |
| 275 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
0.377 |
-0.031 |
| 276 |
249214_at |
AT5G42720
|
[Glycosyl hydrolase family 17 protein] |
0.376 |
0.027 |
| 277 |
249314_at |
AT5G41180
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.376 |
0.121 |
| 278 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.376 |
0.006 |
| 279 |
254632_at |
AT4G18630
|
unknown |
0.375 |
-0.012 |
| 280 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.374 |
0.047 |
| 281 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.374 |
-0.105 |
| 282 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
0.374 |
0.281 |
| 283 |
266311_at |
AT2G27130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.372 |
-0.074 |
| 284 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.371 |
0.048 |
| 285 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.370 |
-0.057 |
| 286 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
0.370 |
0.176 |
| 287 |
260805_at |
AT1G78320
|
ATGSTU23, glutathione S-transferase TAU 23, GSTU23, glutathione S-transferase TAU 23 |
0.370 |
-0.124 |
| 288 |
246505_at |
AT5G16250
|
unknown |
0.369 |
0.223 |
| 289 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.369 |
0.089 |
| 290 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.368 |
0.185 |
| 291 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.368 |
-0.028 |
| 292 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
0.368 |
0.263 |
| 293 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.367 |
-0.109 |
| 294 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.366 |
-0.093 |
| 295 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.365 |
0.142 |
| 296 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.365 |
0.139 |
| 297 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
0.365 |
-0.012 |
| 298 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
0.365 |
0.070 |
| 299 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.364 |
-0.064 |
| 300 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.362 |
0.068 |
| 301 |
248918_at |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
-1.471 |
0.009 |
| 302 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.135 |
-0.099 |
| 303 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-1.082 |
0.293 |
| 304 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.879 |
0.040 |
| 305 |
261198_at |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-0.875 |
0.029 |
| 306 |
246967_at |
AT5G24860
|
ATFPF1, ARABIDOPSIS FLOWERING PROMOTING FACTOR 1, FPF1, FLOWERING PROMOTING FACTOR 1 |
-0.839 |
0.017 |
| 307 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.772 |
-0.073 |
| 308 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.756 |
0.122 |
| 309 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
-0.712 |
0.007 |
| 310 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.699 |
-0.047 |
| 311 |
245086_at |
AT2G39820
|
eIF6B, eukaryotic initiation factor 6B |
-0.695 |
-0.020 |
| 312 |
246997_at |
AT5G67390
|
unknown |
-0.694 |
-0.115 |
| 313 |
262811_at |
AT1G11700
|
unknown |
-0.694 |
-0.197 |
| 314 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
-0.686 |
0.121 |
| 315 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.682 |
-0.032 |
| 316 |
257323_at |
ATMG01200
|
ORF294 |
-0.677 |
0.015 |
| 317 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.658 |
-0.132 |
| 318 |
255957_at |
AT1G22160
|
unknown |
-0.646 |
-0.097 |
| 319 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.643 |
-0.007 |
| 320 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.636 |
-0.013 |
| 321 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.621 |
0.091 |
| 322 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.601 |
-0.198 |
| 323 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.599 |
-0.026 |
| 324 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.594 |
-0.074 |
| 325 |
260149_at |
AT1G52810
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.593 |
-0.110 |
| 326 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.593 |
-0.081 |
| 327 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.592 |
-0.040 |
| 328 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.587 |
0.210 |
| 329 |
247751_at |
AT5G59050
|
unknown |
-0.586 |
-0.105 |
| 330 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.586 |
0.057 |
| 331 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.582 |
-0.007 |
| 332 |
244994_at |
ATCG01010
|
NDHF |
-0.581 |
0.026 |
| 333 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
-0.579 |
-0.109 |
| 334 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.578 |
0.071 |
| 335 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
-0.577 |
0.131 |
| 336 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
-0.575 |
-0.030 |
| 337 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.562 |
-0.109 |
| 338 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.543 |
-0.012 |
| 339 |
249797_at |
AT5G23750
|
[Remorin family protein] |
-0.536 |
-0.080 |
| 340 |
262074_at |
AT1G59670
|
ATGSTU15, glutathione S-transferase TAU 15, GSTU15, glutathione S-transferase TAU 15 |
-0.531 |
0.009 |
| 341 |
262719_at |
AT1G43590
|
unknown |
-0.531 |
0.048 |
| 342 |
246344_at |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
-0.529 |
-0.026 |
| 343 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.525 |
0.021 |
| 344 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.521 |
0.092 |
| 345 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.519 |
0.085 |
| 346 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.517 |
0.402 |
| 347 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
-0.515 |
-0.099 |
| 348 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.513 |
0.016 |
| 349 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-0.509 |
0.113 |
| 350 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.505 |
-0.004 |
| 351 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.504 |
0.491 |
| 352 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.498 |
-0.002 |
| 353 |
260851_at |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-0.495 |
0.018 |
| 354 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.493 |
-0.064 |
| 355 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
-0.490 |
-0.012 |
| 356 |
263292_at |
AT2G10900
|
[expressed protein] |
-0.481 |
-0.067 |
| 357 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
-0.481 |
0.042 |
| 358 |
252888_at |
AT4G39210
|
APL3 |
-0.480 |
-0.058 |
| 359 |
251012_at |
AT5G02580
|
unknown |
-0.478 |
0.091 |
| 360 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.472 |
-0.037 |
| 361 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.459 |
0.013 |
| 362 |
267135_at |
AT2G23430
|
ICK1, KRP1, KIP-RELATED PROTEIN 1 |
-0.459 |
-0.117 |
| 363 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.458 |
-0.050 |
| 364 |
263934_at |
AT2G35950
|
EDA12, embryo sac development arrest 12 |
-0.453 |
-0.043 |
| 365 |
250408_at |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.451 |
-0.188 |
| 366 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.451 |
-0.073 |
| 367 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.448 |
-0.039 |
| 368 |
247051_at |
AT5G66670
|
unknown |
-0.448 |
-0.046 |
| 369 |
255666_at |
AT4G00390
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.447 |
-0.007 |
| 370 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
-0.446 |
-0.010 |
| 371 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.445 |
-0.042 |
| 372 |
264363_at |
AT1G03170
|
FAF2, FANTASTIC FOUR 2, FTM5, FLORAL TRANSITION AT THE MERISTEM5 |
-0.442 |
-0.018 |
| 373 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.436 |
0.065 |
| 374 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.433 |
-0.123 |
| 375 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
-0.433 |
-0.075 |
| 376 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
-0.432 |
-0.049 |
| 377 |
249486_at |
AT5G39030
|
[Protein kinase superfamily protein] |
-0.431 |
-0.036 |
| 378 |
245345_at |
AT4G16640
|
[Matrixin family protein] |
-0.430 |
-0.001 |
| 379 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.427 |
-0.034 |
| 380 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.427 |
0.109 |
| 381 |
250123_at |
AT5G16530
|
PIN5, PIN-FORMED 5 |
-0.426 |
-0.054 |
| 382 |
260567_at |
AT2G43820
|
ATSAGT1, Arabidopsis thaliana salicylic acid glucosyltransferase 1, GT, SAGT1, salicylic acid glucosyltransferase 1, SGT1, UDP-glucose:salicylic acid glucosyltransferase 1, UGT74F2, UDP-glucosyltransferase 74F2 |
-0.426 |
-0.025 |
| 383 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.424 |
-0.028 |
| 384 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.423 |
0.010 |
| 385 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
-0.421 |
-0.073 |
| 386 |
246949_at |
AT5G25140
|
CYP71B13, cytochrome P450, family 71, subfamily B, polypeptide 13 |
-0.420 |
-0.015 |
| 387 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.418 |
-0.009 |
| 388 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.417 |
0.029 |
| 389 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.415 |
-0.014 |
| 390 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-0.412 |
0.068 |
| 391 |
263881_at |
AT2G21820
|
unknown |
-0.411 |
0.010 |
| 392 |
261075_at |
AT1G07280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.411 |
-0.072 |
| 393 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.409 |
-0.011 |
| 394 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
-0.408 |
0.039 |
| 395 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.405 |
0.298 |
| 396 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.403 |
-0.062 |
| 397 |
262582_at |
AT1G15410
|
[aspartate-glutamate racemase family] |
-0.402 |
-0.107 |
| 398 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
-0.401 |
-0.001 |
| 399 |
248114_at |
AT5G55370
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.399 |
0.021 |
| 400 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-0.398 |
-0.056 |
| 401 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
-0.398 |
0.009 |
| 402 |
251207_at |
AT3G63050
|
unknown |
-0.396 |
-0.006 |
| 403 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
-0.395 |
0.006 |
| 404 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-0.391 |
-0.056 |
| 405 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.390 |
-0.167 |
| 406 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.390 |
0.048 |
| 407 |
260926_at |
AT1G21360
|
GLTP2, glycolipid transfer protein 2 |
-0.388 |
-0.005 |
| 408 |
247747_at |
AT5G59000
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.385 |
0.041 |
| 409 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.384 |
0.022 |
| 410 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
-0.384 |
0.036 |
| 411 |
246417_at |
AT5G16990
|
[Zinc-binding dehydrogenase family protein] |
-0.383 |
-0.042 |
| 412 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.382 |
-0.339 |
| 413 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
-0.382 |
-0.028 |
| 414 |
260586_at |
AT2G43630
|
unknown |
-0.382 |
0.071 |
| 415 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-0.381 |
0.023 |
| 416 |
250381_at |
AT5G11610
|
[Exostosin family protein] |
-0.381 |
-0.204 |
| 417 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
-0.381 |
-0.045 |
| 418 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
-0.379 |
-0.059 |
| 419 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.379 |
0.043 |
| 420 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-0.377 |
-0.030 |
| 421 |
264068_at |
AT2G27990
|
BLH8, BEL1-like homeodomain 8, PNF, POUND-FOOLISH |
-0.375 |
-0.027 |
| 422 |
248276_at |
AT5G53550
|
ATYSL3, YELLOW STRIPE LIKE 3, YSL3, YELLOW STRIPE like 3 |
-0.375 |
-0.035 |
| 423 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.375 |
-0.056 |
| 424 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.374 |
-0.053 |
| 425 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
-0.373 |
-0.007 |
| 426 |
263877_at |
AT2G21780
|
unknown |
-0.372 |
-0.014 |
| 427 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.368 |
0.006 |
| 428 |
253662_at |
AT4G30080
|
ARF16, auxin response factor 16 |
-0.368 |
-0.044 |
| 429 |
261456_at |
AT1G21050
|
unknown |
-0.367 |
-0.087 |
| 430 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.367 |
-0.080 |
| 431 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.362 |
0.113 |
| 432 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.361 |
0.146 |
| 433 |
263297_at |
AT2G15310
|
ARFB1A, ADP-ribosylation factor B1A, ATARFB1A, ADP-ribosylation factor B1A |
-0.361 |
-0.068 |
| 434 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.360 |
-0.064 |
| 435 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.359 |
0.038 |
| 436 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.358 |
0.020 |
| 437 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
-0.357 |
0.128 |
| 438 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
-0.357 |
-0.010 |
| 439 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.357 |
-0.043 |
| 440 |
266719_at |
AT2G46830
|
AtCCA1, CCA1, circadian clock associated 1 |
-0.357 |
-0.314 |
| 441 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.356 |
0.002 |
| 442 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.356 |
0.015 |
| 443 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
-0.356 |
0.047 |
| 444 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.356 |
-0.129 |
| 445 |
252319_at |
AT3G48710
|
[DEK domain-containing chromatin associated protein] |
-0.356 |
0.042 |
| 446 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.353 |
-0.229 |
| 447 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-0.353 |
-0.055 |
| 448 |
258203_at |
AT3G13950
|
unknown |
-0.352 |
0.113 |
| 449 |
251983_at |
AT3G53210
|
UMAMIT6, Usually multiple acids move in and out Transporters 6 |
-0.352 |
0.084 |
| 450 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
-0.351 |
-0.059 |
| 451 |
254723_at |
AT4G13510
|
AMT1;1, ammonium transporter 1;1, ATAMT1, ARABIDOPSIS THALIANA AMMONIUM TRANSPORT 1, ATAMT1;1 |
-0.351 |
-0.004 |
| 452 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.350 |
-0.069 |
| 453 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.350 |
0.031 |
| 454 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.350 |
0.084 |
| 455 |
260005_at |
AT1G67920
|
unknown |
-0.349 |
0.110 |
| 456 |
264968_at |
AT1G67360
|
[Rubber elongation factor protein (REF)] |
-0.349 |
0.252 |
| 457 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
-0.348 |
0.001 |
| 458 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
-0.348 |
0.128 |
| 459 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.347 |
0.026 |
| 460 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.346 |
-0.063 |
| 461 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
-0.345 |
-0.103 |
| 462 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
-0.345 |
0.089 |
| 463 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.341 |
-0.022 |
| 464 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.341 |
0.016 |
| 465 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
-0.341 |
0.000 |
| 466 |
265611_at |
AT2G25510
|
unknown |
-0.341 |
-0.016 |
| 467 |
259436_at |
AT1G01500
|
[Erythronate-4-phosphate dehydrogenase family protein] |
-0.340 |
-0.066 |
| 468 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.340 |
-0.111 |
| 469 |
262421_at |
AT1G50290
|
unknown |
-0.339 |
-0.117 |
| 470 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.338 |
-0.031 |
| 471 |
260156_at |
AT1G52880
|
ANAC018, Arabidopsis NAC domain containing protein 18, ATNAM, NAM, NO APICAL MERISTEM, NARS2, NAC-REGULATED SEED MORPHOLOGY 2 |
-0.338 |
-0.153 |
| 472 |
258807_at |
AT3G04030
|
MYR2 |
-0.337 |
-0.167 |
| 473 |
265937_at |
AT2G19610
|
[RING/U-box superfamily protein] |
-0.337 |
-0.056 |
| 474 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
-0.337 |
0.011 |
| 475 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.336 |
0.310 |
| 476 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.335 |
-0.170 |
| 477 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.334 |
0.060 |
| 478 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.333 |
-0.054 |
| 479 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.332 |
0.531 |
| 480 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.332 |
0.057 |
| 481 |
244938_at |
ATCG01120
|
RPS15, chloroplast ribosomal protein S15 |
-0.331 |
-0.026 |
| 482 |
255403_at |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-0.331 |
0.006 |
| 483 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.331 |
0.028 |
| 484 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.328 |
-0.079 |
| 485 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.328 |
0.122 |
| 486 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.327 |
0.004 |
| 487 |
259520_at |
AT1G12320
|
unknown |
-0.327 |
0.065 |
| 488 |
258103_at |
AT3G23630
|
ATIPT7, ARABIDOPSIS THALIANA ISOPENTENYLTRANSFERASE 7, IPT7, isopentenyltransferase 7 |
-0.326 |
-0.046 |
| 489 |
254752_at |
AT4G13160
|
unknown |
-0.325 |
-0.061 |
| 490 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.325 |
-0.017 |
| 491 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.325 |
0.009 |
| 492 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
-0.322 |
0.012 |
| 493 |
246133_at |
AT5G20960
|
AAO1, aldehyde oxidase 1, AO1, aldehyde oxidase 1, AOalpha, aldehyde oxidase alpha, AT-AO1, ARABIDOPSIS THALIANA ALDEHYDE OXIDASE 1, ATAO, AtAO1, Arabidopsis thaliana aldehyde oxidase 1 |
-0.322 |
0.135 |
| 494 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
-0.322 |
0.014 |
| 495 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
-0.321 |
0.048 |
| 496 |
251743_at |
AT3G55890
|
[Yippee family putative zinc-binding protein] |
-0.320 |
0.014 |
| 497 |
262870_at |
AT1G64710
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.319 |
-0.028 |
| 498 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
-0.319 |
0.028 |
| 499 |
253940_at |
AT4G26950
|
unknown |
-0.318 |
-0.059 |
| 500 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.318 |
-0.062 |
| 501 |
246222_at |
AT4G36900
|
DEAR4, DREB AND EAR MOTIF PROTEIN 4, RAP2.10, related to AP2 10 |
-0.317 |
0.064 |
| 502 |
253827_at |
AT4G28085
|
unknown |
-0.317 |
0.029 |
| 503 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.317 |
-0.073 |
| 504 |
255835_at |
AT2G33420
|
unknown |
-0.316 |
-0.012 |
| 505 |
257946_at |
AT3G21710
|
unknown |
-0.315 |
-0.072 |
| 506 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.315 |
-0.061 |
| 507 |
251254_at |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
-0.314 |
-0.044 |
| 508 |
245055_at |
AT2G26470
|
unknown |
-0.314 |
-0.044 |
| 509 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.314 |
0.137 |
| 510 |
260420_at |
AT1G69610
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.314 |
-0.021 |
| 511 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
-0.312 |
-0.061 |
| 512 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
-0.312 |
0.047 |
| 513 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.311 |
0.092 |
| 514 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
-0.311 |
0.031 |
| 515 |
261073_at |
AT1G07300
|
[josephin protein-related] |
-0.311 |
-0.033 |
| 516 |
246989_at |
AT5G67350
|
unknown |
-0.310 |
-0.031 |
| 517 |
253956_at |
AT4G26700
|
ATFIM1, ARABIDOPSIS THALIANA FIMBRIN 1, FIM1, fimbrin 1 |
-0.310 |
-0.043 |
| 518 |
257872_at |
AT3G28360
|
ABCB16, ATP-binding cassette B16, PGP16, P-glycoprotein 16 |
-0.309 |
-0.036 |
| 519 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.309 |
-0.013 |
| 520 |
263283_at |
AT2G36090
|
[F-box family protein] |
-0.308 |
-0.027 |
| 521 |
245354_at |
AT4G17600
|
LIL3:1 |
-0.307 |
-0.018 |
| 522 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.307 |
0.037 |
| 523 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.306 |
0.018 |
| 524 |
245544_at |
AT4G15270
|
[glucosyltransferase-related] |
-0.306 |
-0.046 |
| 525 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.305 |
0.023 |
| 526 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.305 |
0.017 |
| 527 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.305 |
-0.023 |
| 528 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
-0.304 |
0.027 |
| 529 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.303 |
-0.025 |
| 530 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
-0.303 |
-0.096 |
| 531 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
-0.303 |
-0.047 |
| 532 |
256422_at |
AT1G33520
|
MOS2, modifier of snc1, 2 |
-0.303 |
-0.019 |
| 533 |
246209_at |
AT4G36870
|
BLH2, BEL1-like homeodomain 2, SAW1, SAWTOOTH 1 |
-0.303 |
0.008 |
| 534 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
-0.302 |
0.073 |
| 535 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
-0.302 |
0.007 |
| 536 |
261894_at |
AT1G80900
|
ATMGT1, MAGNESIUM TRANSPORTER 1, MGT1, magnesium transporter 1, MRS2-10 |
-0.301 |
-0.078 |
| 537 |
246783_at |
AT5G27360
|
SFP2 |
-0.301 |
-0.073 |
| 538 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.301 |
-0.045 |
| 539 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.300 |
0.058 |
| 540 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.299 |
0.411 |
| 541 |
265908_at |
AT4G00270
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.298 |
0.035 |
| 542 |
256436_at |
AT3G11150
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.298 |
-0.007 |
| 543 |
254921_at |
AT4G11300
|
unknown |
-0.297 |
0.087 |
| 544 |
253793_at |
AT4G28710
|
ATXIH, MYOSIN XI H, XIH |
-0.297 |
0.014 |
| 545 |
247998_at |
AT5G56200
|
[C2H2 type zinc finger transcription factor family] |
-0.296 |
-0.011 |
| 546 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.296 |
-0.003 |
| 547 |
254574_at |
AT4G19430
|
unknown |
-0.295 |
0.267 |
| 548 |
267096_at |
AT2G38180
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.295 |
-0.036 |
| 549 |
259372_at |
AT1G69120
|
AGL7, AGAMOUS-like 7, AP1, APETALA1, AtAP1 |
-0.294 |
-0.032 |
| 550 |
261300_at |
AT1G48560
|
unknown |
-0.294 |
0.060 |
| 551 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.293 |
-0.001 |
| 552 |
263225_at |
AT1G30650
|
AR411, ATWRKY14, WRKY DNA-BINDING PROTEIN 14, WRKY14, WRKY DNA-binding protein 14 |
-0.293 |
-0.068 |
| 553 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.292 |
-0.096 |
| 554 |
265893_at |
AT2G15042
|
[Leucine-rich repeat (LRR) family protein] |
-0.292 |
0.018 |
| 555 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.292 |
-0.007 |
| 556 |
248889_at |
AT5G46230
|
unknown |
-0.292 |
-0.197 |
| 557 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.292 |
-0.035 |
| 558 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
-0.291 |
-0.117 |
| 559 |
255551_at |
AT4G01840
|
ATKCO5, ATTPK5, KCO5, Ca2+ activated outward rectifying K+ channel 5, TPK5 |
-0.291 |
-0.094 |
| 560 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-0.291 |
0.007 |
| 561 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.290 |
0.002 |
| 562 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
-0.289 |
0.012 |
| 563 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.289 |
0.142 |
| 564 |
253155_at |
AT4G35720
|
unknown |
-0.288 |
-0.008 |
| 565 |
246796_at |
AT5G26770
|
unknown |
-0.288 |
0.206 |
| 566 |
256764_at |
AT3G29310
|
[calmodulin-binding protein-related] |
-0.287 |
0.042 |
| 567 |
250948_at |
AT5G03490
|
[UDP-Glycosyltransferase superfamily protein] |
-0.287 |
-0.105 |
| 568 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.287 |
0.120 |
| 569 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.286 |
0.025 |
| 570 |
247874_at |
AT5G57710
|
SMAX1, SUPPRESSOR OF MAX2 1 |
-0.285 |
-0.011 |
| 571 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
-0.285 |
-0.099 |
| 572 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.285 |
0.094 |
| 573 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.285 |
-0.023 |
| 574 |
255903_at |
AT1G17950
|
ATMYB52, MYB DOMAIN PROTEIN 52, BW52, MYB52, myb domain protein 52 |
-0.285 |
0.016 |
| 575 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.284 |
-0.121 |
| 576 |
267021_at |
AT2G39300
|
unknown |
-0.284 |
0.028 |
| 577 |
249357_at |
AT5G40490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.284 |
-0.016 |
| 578 |
266824_at |
AT2G22800
|
HAT9 |
-0.284 |
-0.090 |
| 579 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.283 |
0.034 |
| 580 |
260189_at |
AT1G67550
|
URE, urease |
-0.283 |
-0.031 |
| 581 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.283 |
-0.159 |
| 582 |
263611_at |
AT2G16450
|
[F-box and associated interaction domains-containing protein] |
-0.283 |
-0.015 |
| 583 |
250906_at |
AT5G03650
|
SBE2.2, starch branching enzyme 2.2 |
-0.283 |
-0.029 |
| 584 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
-0.282 |
-0.048 |
| 585 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.282 |
0.151 |
| 586 |
263787_at |
AT2G46420
|
unknown |
-0.282 |
-0.038 |
| 587 |
247503_at |
AT5G61980
|
AGD1, ARF-GAP domain 1 |
-0.282 |
0.008 |
| 588 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.281 |
-0.068 |
| 589 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
-0.281 |
-0.015 |
| 590 |
251221_at |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.281 |
0.023 |
| 591 |
266695_at |
AT2G19810
|
AtOZF1, AtTZF2, OZF1, Oxidation-related Zinc Finger 1, TZF2, tandem zinc finger 2 |
-0.280 |
0.001 |
| 592 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
-0.279 |
0.251 |
| 593 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
-0.279 |
0.105 |
| 594 |
263569_at |
AT2G27170
|
SMC3, STRUCTURAL MAINTENANCE OF CHROMOSOMES 3, TTN7, TITAN7 |
-0.279 |
0.038 |
| 595 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
-0.278 |
0.024 |
| 596 |
255028_at |
AT4G09890
|
unknown |
-0.276 |
-0.008 |
| 597 |
250722_at |
AT5G06190
|
unknown |
-0.276 |
-0.016 |
| 598 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.276 |
0.090 |
| 599 |
248037_at |
AT5G55930
|
ATOPT1, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 1, OPT1, oligopeptide transporter 1 |
-0.276 |
0.031 |
| 600 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.275 |
-0.078 |