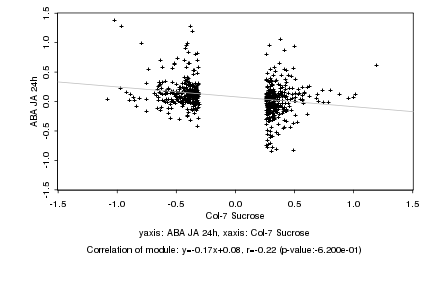
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Col-7 Sucrose |
Log2 signal ratio
ABA JA 24h |
| 1 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.189 |
0.622 |
| 2 |
252888_at |
AT4G39210
|
APL3 |
1.009 |
0.120 |
| 3 |
256139_at |
AT1G48660
|
[Auxin-responsive GH3 family protein] |
0.994 |
0.085 |
| 4 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.954 |
0.062 |
| 5 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.877 |
0.132 |
| 6 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.800 |
0.204 |
| 7 |
256924_at |
AT3G29590
|
AT5MAT |
0.780 |
-0.000 |
| 8 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.741 |
-0.003 |
| 9 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
0.734 |
0.192 |
| 10 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.697 |
0.014 |
| 11 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.693 |
0.134 |
| 12 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.678 |
0.067 |
| 13 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
0.622 |
0.272 |
| 14 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.622 |
0.097 |
| 15 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.606 |
0.248 |
| 16 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.605 |
-0.215 |
| 17 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.579 |
0.152 |
| 18 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.577 |
0.162 |
| 19 |
264215_at |
AT1G65340
|
CYP96A3, cytochrome P450, family 96, subfamily A, polypeptide 3 |
0.571 |
-0.029 |
| 20 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.566 |
0.249 |
| 21 |
252988_at |
AT4G38410
|
[Dehydrin family protein] |
0.563 |
0.109 |
| 22 |
249813_at |
AT5G23940
|
DCR, DEFECTIVE IN CUTICULAR RIDGES, EMB3009, EMBRYO DEFECTIVE 3009, PEL3, PERMEABLE LEAVES3 |
0.559 |
0.042 |
| 23 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.559 |
0.024 |
| 24 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
0.546 |
0.045 |
| 25 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.541 |
0.147 |
| 26 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
0.533 |
0.121 |
| 27 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.533 |
0.209 |
| 28 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
0.521 |
-0.350 |
| 29 |
246797_at |
AT5G26790
|
unknown |
0.508 |
-0.229 |
| 30 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.508 |
0.383 |
| 31 |
250558_at |
AT5G07990
|
CYP75B1, CYTOCHROME P450 75B1, D501, TT7, TRANSPARENT TESTA 7 |
0.505 |
0.140 |
| 32 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.505 |
0.017 |
| 33 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.505 |
0.079 |
| 34 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.498 |
0.936 |
| 35 |
263829_at |
AT2G40435
|
unknown |
0.492 |
0.010 |
| 36 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.485 |
-0.357 |
| 37 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.483 |
0.566 |
| 38 |
252763_at |
AT3G42725
|
[Putative membrane lipoprotein] |
0.483 |
-0.827 |
| 39 |
259353_at |
AT3G05190
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.481 |
0.053 |
| 40 |
262347_at |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
0.478 |
0.128 |
| 41 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.475 |
0.223 |
| 42 |
257724_at |
AT3G18510
|
unknown |
0.472 |
-0.137 |
| 43 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.468 |
0.433 |
| 44 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
0.465 |
0.042 |
| 45 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.460 |
-0.431 |
| 46 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.454 |
-0.077 |
| 47 |
251317_at |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
0.453 |
0.233 |
| 48 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.446 |
0.458 |
| 49 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.444 |
-0.024 |
| 50 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.442 |
0.138 |
| 51 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.434 |
0.036 |
| 52 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.430 |
0.537 |
| 53 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.429 |
0.090 |
| 54 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.428 |
-0.328 |
| 55 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.427 |
-0.129 |
| 56 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.427 |
0.325 |
| 57 |
250517_at |
AT5G08260
|
scpl35, serine carboxypeptidase-like 35 |
0.426 |
-0.155 |
| 58 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
0.424 |
-0.146 |
| 59 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
0.422 |
0.076 |
| 60 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.421 |
0.387 |
| 61 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.421 |
-0.019 |
| 62 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.417 |
-0.157 |
| 63 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.417 |
0.205 |
| 64 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.416 |
0.278 |
| 65 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.415 |
-0.429 |
| 66 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.413 |
0.556 |
| 67 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.412 |
0.871 |
| 68 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.411 |
0.156 |
| 69 |
246249_at |
AT4G36680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.410 |
-0.095 |
| 70 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.410 |
0.108 |
| 71 |
256675_at |
AT3G52170
|
[DNA binding] |
0.408 |
-0.312 |
| 72 |
252361_at |
AT3G48490
|
unknown |
0.408 |
-0.148 |
| 73 |
251005_at |
AT5G02590
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.406 |
-0.134 |
| 74 |
263535_at |
AT2G24970
|
unknown |
0.405 |
-0.454 |
| 75 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.404 |
-0.164 |
| 76 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.398 |
0.164 |
| 77 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
0.397 |
0.112 |
| 78 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.394 |
0.241 |
| 79 |
261921_at |
AT1G65900
|
unknown |
0.394 |
0.092 |
| 80 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.393 |
-0.193 |
| 81 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
0.393 |
-0.029 |
| 82 |
262103_at |
AT1G02940
|
ATGSTF5, GLUTATHIONE S-TRANSFERASE (CLASS PHI) 5, GSTF5, glutathione S-transferase (class phi) 5 |
0.391 |
-0.091 |
| 83 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.390 |
0.021 |
| 84 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
0.387 |
0.136 |
| 85 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
0.386 |
0.448 |
| 86 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.380 |
1.059 |
| 87 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.379 |
0.186 |
| 88 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.377 |
0.121 |
| 89 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
0.375 |
0.137 |
| 90 |
263730_at |
AT1G60090
|
BGLU4, beta glucosidase 4 |
0.374 |
0.228 |
| 91 |
259928_at |
AT1G34380
|
[5'-3' exonuclease family protein] |
0.374 |
-0.081 |
| 92 |
254208_at |
AT4G24175
|
unknown |
0.373 |
-0.058 |
| 93 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.373 |
0.276 |
| 94 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.372 |
0.110 |
| 95 |
261305_at |
AT1G48470
|
GLN1;5, glutamine synthetase 1;5 |
0.371 |
0.072 |
| 96 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
0.370 |
-0.204 |
| 97 |
255527_at |
AT4G02360
|
unknown |
0.365 |
0.648 |
| 98 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
0.365 |
0.019 |
| 99 |
251685_at |
AT3G56430
|
unknown |
0.364 |
-0.134 |
| 100 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.362 |
-0.263 |
| 101 |
246786_at |
AT5G27410
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.359 |
0.110 |
| 102 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.358 |
-0.544 |
| 103 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.358 |
0.148 |
| 104 |
247024_at |
AT5G66985
|
unknown |
0.357 |
-0.025 |
| 105 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.357 |
-0.374 |
| 106 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.357 |
-0.099 |
| 107 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
0.356 |
0.290 |
| 108 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
0.350 |
-0.062 |
| 109 |
249756_at |
AT5G24313
|
unknown |
0.350 |
0.036 |
| 110 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
0.349 |
0.042 |
| 111 |
246002_at |
AT5G20740
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.348 |
0.020 |
| 112 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.348 |
0.100 |
| 113 |
263679_at |
AT1G59990
|
EMB3108, EMBRYO DEFECTIVE 3108, HS3, HEAVY SEED 3, RH22, RNA helicase 22 |
0.348 |
-0.175 |
| 114 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.346 |
-0.170 |
| 115 |
248596_at |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
0.343 |
0.086 |
| 116 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.342 |
0.372 |
| 117 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.341 |
-0.088 |
| 118 |
258471_at |
AT3G06030
|
ANP3, NPK1-related protein kinase 3, AtANP3, MAPKKK12, NP3, NPK1-related protein kinase 3 |
0.340 |
-0.526 |
| 119 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.340 |
-0.127 |
| 120 |
246415_at |
AT5G17160
|
unknown |
0.339 |
-0.800 |
| 121 |
258367_at |
AT3G14370
|
WAG2 |
0.338 |
0.093 |
| 122 |
260605_at |
AT2G43780
|
unknown |
0.338 |
-0.052 |
| 123 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.337 |
0.341 |
| 124 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.336 |
0.134 |
| 125 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
0.333 |
-0.111 |
| 126 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.333 |
0.516 |
| 127 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.332 |
-0.142 |
| 128 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.332 |
-0.052 |
| 129 |
256858_at |
AT3G15140
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.331 |
-0.151 |
| 130 |
265596_at |
AT2G20020
|
ATCAF1, CAF1 |
0.331 |
-0.043 |
| 131 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.330 |
-0.221 |
| 132 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.329 |
-0.063 |
| 133 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
0.329 |
0.178 |
| 134 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.329 |
0.064 |
| 135 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.329 |
0.100 |
| 136 |
253917_at |
AT4G27380
|
unknown |
0.327 |
-0.070 |
| 137 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
0.327 |
0.051 |
| 138 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
0.326 |
-0.040 |
| 139 |
266264_at |
AT2G27775
|
unknown |
0.326 |
-0.277 |
| 140 |
245163_at |
AT2G33230
|
YUC7, YUCCA 7 |
0.325 |
0.048 |
| 141 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
0.325 |
0.011 |
| 142 |
250603_at |
AT5G07820
|
[Plant calmodulin-binding protein-related] |
0.325 |
-0.142 |
| 143 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
0.325 |
-0.299 |
| 144 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.324 |
0.587 |
| 145 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.324 |
-0.225 |
| 146 |
261502_at |
AT1G14440
|
AtHB31, homeobox protein 31, FTM2, FLORAL TRANSITION AT THE MERISTEM2, HB31, homeobox protein 31, ZHD4, ZINC FINGER HOMEODOMAIN 4 |
0.323 |
0.128 |
| 147 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
0.323 |
-0.031 |
| 148 |
249015_at |
AT5G44730
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.322 |
0.079 |
| 149 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.321 |
0.106 |
| 150 |
249826_at |
AT5G23310
|
FSD3, Fe superoxide dismutase 3 |
0.320 |
0.008 |
| 151 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
0.320 |
-0.050 |
| 152 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
0.320 |
0.107 |
| 153 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.319 |
0.459 |
| 154 |
251660_at |
AT3G57160
|
unknown |
0.318 |
-0.168 |
| 155 |
252995_at |
AT4G38370
|
[Phosphoglycerate mutase family protein] |
0.318 |
0.076 |
| 156 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.317 |
-0.271 |
| 157 |
250152_at |
AT5G15120
|
unknown |
0.317 |
0.058 |
| 158 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.316 |
0.215 |
| 159 |
249522_at |
AT5G38700
|
unknown |
0.315 |
-0.242 |
| 160 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.315 |
-0.258 |
| 161 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.315 |
-0.190 |
| 162 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.315 |
0.170 |
| 163 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.315 |
0.125 |
| 164 |
257213_at |
AT3G15020
|
mMDH2, mitochondrial malate dehydrogenase 2 |
0.314 |
0.083 |
| 165 |
255224_at |
AT4G05400
|
[copper ion binding] |
0.314 |
-0.102 |
| 166 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.312 |
-0.020 |
| 167 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
0.311 |
-0.271 |
| 168 |
264133_at |
AT1G79080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.311 |
-0.241 |
| 169 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.310 |
0.108 |
| 170 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.310 |
0.178 |
| 171 |
258867_at |
AT3G03130
|
unknown |
0.309 |
-0.306 |
| 172 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.309 |
0.069 |
| 173 |
256832_at |
AT3G22880
|
ARLIM15, ARABIDOPSIS HOMOLOG OF LILY MESSAGES INDUCED AT MEIOSIS 15, ATDMC1, ARABIDOPSIS THALIANA DISRUPTION OF MEIOTIC CONTROL 1, DMC1, DISRUPTION OF MEIOTIC CONTROL 1 |
0.309 |
-0.022 |
| 174 |
264763_at |
AT1G61450
|
unknown |
0.308 |
-0.767 |
| 175 |
253008_at |
AT4G38210
|
ATEXP20, ATEXPA20, expansin A20, ATHEXP ALPHA 1.23, EXP20, EXPANSIN 20, EXPA20, expansin A20 |
0.308 |
-0.090 |
| 176 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.308 |
0.149 |
| 177 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.307 |
0.274 |
| 178 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.307 |
0.064 |
| 179 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.307 |
-0.086 |
| 180 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.306 |
-0.196 |
| 181 |
259366_at |
AT1G13280
|
AOC4, allene oxide cyclase 4 |
0.306 |
0.294 |
| 182 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
0.306 |
-0.578 |
| 183 |
254932_at |
AT4G11120
|
[translation elongation factor Ts (EF-Ts), putative] |
0.306 |
-0.043 |
| 184 |
261247_at |
AT1G20070
|
unknown |
0.304 |
0.156 |
| 185 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.303 |
0.361 |
| 186 |
247425_at |
AT5G62550
|
unknown |
0.303 |
-0.834 |
| 187 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.303 |
0.349 |
| 188 |
245586_at |
AT4G14980
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.302 |
0.349 |
| 189 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.302 |
-0.425 |
| 190 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.299 |
-0.073 |
| 191 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.299 |
0.291 |
| 192 |
257186_at |
AT3G13130
|
unknown |
0.299 |
0.078 |
| 193 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.298 |
0.183 |
| 194 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
0.298 |
-0.018 |
| 195 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.298 |
-0.068 |
| 196 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.296 |
-0.149 |
| 197 |
254132_at |
AT4G24660
|
ATHB22, HOMEOBOX PROTEIN 22, HB22, homeobox protein 22, MEE68, MATERNAL EFFECT EMBRYO ARREST 68, ZHD2, ZINC FINGER HOMEODOMAIN 2 |
0.296 |
0.119 |
| 198 |
256409_at |
AT1G66620
|
[Protein with RING/U-box and TRAF-like domains] |
0.296 |
-0.199 |
| 199 |
248057_at |
AT5G55520
|
unknown |
0.296 |
-0.704 |
| 200 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.296 |
0.265 |
| 201 |
263096_at |
AT2G16060
|
AHB1, hemoglobin 1, ARATH GLB1, ATGLB1, GLB1, CLASS I HEMOGLOBIN, HB1, hemoglobin 1, NSHB1 |
0.296 |
-0.112 |
| 202 |
252097_at |
AT3G51090
|
unknown |
0.296 |
0.398 |
| 203 |
258376_at |
AT3G17680
|
[Kinase interacting (KIP1-like) family protein] |
0.295 |
-0.576 |
| 204 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.295 |
0.109 |
| 205 |
251535_at |
AT3G58540
|
unknown |
0.294 |
-0.101 |
| 206 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.294 |
-0.497 |
| 207 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.293 |
-0.024 |
| 208 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.293 |
-0.607 |
| 209 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
0.293 |
0.256 |
| 210 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.292 |
0.366 |
| 211 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.292 |
0.038 |
| 212 |
258316_at |
AT3G22660
|
[rRNA processing protein-related] |
0.292 |
-0.175 |
| 213 |
252103_at |
AT3G51410
|
unknown |
0.291 |
0.065 |
| 214 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
0.290 |
0.555 |
| 215 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.290 |
-0.312 |
| 216 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.289 |
-0.050 |
| 217 |
256114_at |
AT1G16850
|
unknown |
0.289 |
0.220 |
| 218 |
258555_at |
AT3G06860
|
ATMFP2, MULTIFUNCTIONAL PROTEIN 2, MFP2, multifunctional protein 2 |
0.289 |
0.196 |
| 219 |
262659_at |
AT1G14240
|
[GDA1/CD39 nucleoside phosphatase family protein] |
0.288 |
0.964 |
| 220 |
255360_at |
AT4G03960
|
AtPFA-DSP4, PFA-DSP4, plant and fungi atypical dual-specificity phosphatase 4 |
0.288 |
0.339 |
| 221 |
259985_at |
AT1G76620
|
unknown |
0.287 |
0.151 |
| 222 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.287 |
-0.078 |
| 223 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
0.287 |
0.061 |
| 224 |
260028_at |
AT1G29980
|
unknown |
0.285 |
-0.269 |
| 225 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
0.285 |
-0.164 |
| 226 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.285 |
-0.283 |
| 227 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.284 |
0.192 |
| 228 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.284 |
0.073 |
| 229 |
245937_at |
AT5G19750
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.283 |
-0.161 |
| 230 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.282 |
-0.328 |
| 231 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.282 |
0.094 |
| 232 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.282 |
0.120 |
| 233 |
263544_at |
AT2G21590
|
APL4 |
0.282 |
0.098 |
| 234 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.281 |
0.078 |
| 235 |
251025_at |
AT5G02190
|
ATASP38, ARABIDOPSIS THALIANA ASPARTIC PROTEASE 38, EMB24, EMBRYO DEFECTIVE 24, PCS1, PROMOTION OF CELL SURVIVAL 1 |
0.281 |
0.021 |
| 236 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
0.280 |
0.109 |
| 237 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
0.280 |
-0.033 |
| 238 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
0.279 |
0.137 |
| 239 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.279 |
-0.724 |
| 240 |
247904_at |
AT5G57390
|
AIL5, AINTEGUMENTA-like 5, CHO1, CHOTTO 1, EMK, EMBRYOMAKER, PLT5, PLETHORA 5 |
0.278 |
0.353 |
| 241 |
262706_at |
AT1G16280
|
AtRH36, Arabidopsis thaliana RNA helicase 36, RH36, RNA helicase 36, SWA3, SLOW WALKER 3 |
0.278 |
-0.077 |
| 242 |
266801_at |
AT2G22870
|
EMB2001, embryo defective 2001 |
0.278 |
-0.129 |
| 243 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.278 |
0.064 |
| 244 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.278 |
-0.213 |
| 245 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.277 |
-0.021 |
| 246 |
250679_at |
AT5G06550
|
JMJ22, Jumonji domain-containing protein 22 |
0.276 |
-0.337 |
| 247 |
248341_at |
AT5G52220
|
unknown |
0.276 |
-0.302 |
| 248 |
249347_at |
AT5G40830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.276 |
-0.325 |
| 249 |
249167_at |
AT5G42860
|
unknown |
0.276 |
-0.201 |
| 250 |
265948_at |
AT2G19590
|
ACO1, ACC oxidase 1, ATACO1 |
0.274 |
0.130 |
| 251 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.273 |
0.197 |
| 252 |
251278_at |
AT3G61780
|
emb1703, embryo defective 1703 |
0.272 |
-0.105 |
| 253 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.272 |
0.100 |
| 254 |
260462_at |
AT1G10970
|
ATZIP4, ZIP4, zinc transporter 4 precursor |
0.272 |
0.138 |
| 255 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.271 |
-0.282 |
| 256 |
260292_at |
AT1G63680
|
APG13, ALBINO OR PALE-GREEN 13, ATMURE, MURE, PDE316, PIGMENT DEFECTIVE EMBRYO 316 |
0.271 |
-0.043 |
| 257 |
253020_at |
AT4G38020
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
0.271 |
0.111 |
| 258 |
265442_at |
AT2G20940
|
unknown |
0.271 |
-0.054 |
| 259 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
0.270 |
-0.655 |
| 260 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
0.270 |
-0.273 |
| 261 |
255460_at |
AT4G02800
|
unknown |
0.270 |
-0.774 |
| 262 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.270 |
0.144 |
| 263 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.269 |
-0.056 |
| 264 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.269 |
-0.174 |
| 265 |
253804_at |
AT4G28230
|
unknown |
0.268 |
-0.477 |
| 266 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.268 |
-0.204 |
| 267 |
255900_at |
AT1G17830
|
unknown |
0.267 |
0.482 |
| 268 |
266423_at |
AT2G41340
|
RPB5D, RNA polymerase II fifth largest subunit, D |
0.267 |
0.025 |
| 269 |
262875_at |
AT1G64970
|
G-TMT, gamma-tocopherol methyltransferase, TMT1, VTE4, VITAMIN E DEFICIENT 4 |
0.267 |
0.255 |
| 270 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.267 |
-0.558 |
| 271 |
256043_at |
AT1G07210
|
[Ribosomal protein S18] |
0.266 |
0.053 |
| 272 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
0.266 |
0.085 |
| 273 |
247329_at |
AT5G64150
|
[RNA methyltransferase family protein] |
0.266 |
-0.043 |
| 274 |
266206_at |
AT2G27730
|
[copper ion binding] |
0.265 |
-0.051 |
| 275 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.265 |
-0.081 |
| 276 |
262880_at |
AT1G64880
|
[Ribosomal protein S5 family protein] |
0.265 |
-0.152 |
| 277 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.265 |
-0.087 |
| 278 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.264 |
0.267 |
| 279 |
257970_at |
AT3G27570
|
[Sucrase/ferredoxin-like family protein] |
0.264 |
0.065 |
| 280 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.263 |
0.071 |
| 281 |
262937_at |
AT1G79560
|
EMB1047, EMBRYO DEFECTIVE 1047, EMB156, EMBRYO DEFECTIVE 156, EMB36, EMBRYO DEFECTIVE 36, FTSH12, FTSH protease 12 |
0.263 |
-0.052 |
| 282 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.262 |
0.134 |
| 283 |
251090_at |
AT5G01340
|
AtmSFC1, mSFC1, mitochondrial succinate-fumarate carrier 1 |
0.262 |
-0.026 |
| 284 |
264806_at |
AT1G08610
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.262 |
-0.215 |
| 285 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
0.261 |
0.088 |
| 286 |
253978_at |
AT4G26660
|
unknown |
0.260 |
-0.732 |
| 287 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.260 |
-0.149 |
| 288 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.260 |
0.377 |
| 289 |
260682_at |
AT1G17510
|
unknown |
0.260 |
0.031 |
| 290 |
246057_at |
AT5G08400
|
unknown |
0.260 |
0.037 |
| 291 |
265011_at |
AT1G24490
|
ALB4, ALBINA 4, ARTEMIS, ARABIDOPSIS THALIANA ENVELOPE MEMBRANE INTEGRASE |
0.260 |
0.058 |
| 292 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.259 |
0.015 |
| 293 |
261043_at |
AT1G01220
|
AtFKGP, Arabidopsis thaliana L-fucokinase/GDP-L-fucose pyrophosphorylase, FKGP, L-fucokinase/GDP-L-fucose pyrophosphorylase |
0.259 |
0.075 |
| 294 |
246767_at |
AT5G27395
|
[Mitochondrial inner membrane translocase complex, subunit Tim44-related protein] |
0.259 |
-0.301 |
| 295 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.258 |
0.374 |
| 296 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
0.258 |
-0.381 |
| 297 |
255032_at |
AT4G09500
|
[UDP-Glycosyltransferase superfamily protein] |
0.258 |
0.801 |
| 298 |
255487_at |
AT4G02610
|
[Aldolase-type TIM barrel family protein] |
0.258 |
0.054 |
| 299 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.258 |
-0.302 |
| 300 |
260742_at |
AT1G15050
|
IAA34, indole-3-acetic acid inducible 34 |
0.257 |
0.143 |
| 301 |
256527_at |
AT1G66100
|
[Plant thionin] |
-1.083 |
0.047 |
| 302 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-1.026 |
1.393 |
| 303 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.976 |
0.236 |
| 304 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.964 |
1.280 |
| 305 |
259765_at |
AT1G64370
|
unknown |
-0.923 |
0.154 |
| 306 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.901 |
0.034 |
| 307 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.890 |
0.127 |
| 308 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.870 |
0.075 |
| 309 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.858 |
0.021 |
| 310 |
245925_at |
AT5G28770
|
AtbZIP63, Arabidopsis thaliana basic leucine zipper 63, BZO2H3 |
-0.840 |
-0.076 |
| 311 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.813 |
0.057 |
| 312 |
265387_at |
AT2G20670
|
unknown |
-0.795 |
0.997 |
| 313 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.757 |
0.319 |
| 314 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.756 |
0.046 |
| 315 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.756 |
-0.153 |
| 316 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.738 |
0.556 |
| 317 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.689 |
0.142 |
| 318 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.671 |
0.107 |
| 319 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.667 |
-0.102 |
| 320 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.662 |
0.206 |
| 321 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.658 |
0.074 |
| 322 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.657 |
0.261 |
| 323 |
258262_at |
AT3G15770
|
unknown |
-0.656 |
0.026 |
| 324 |
251356_at |
AT3G61060
|
AtPP2-A13, phloem protein 2-A13, PP2-A13, phloem protein 2-A13 |
-0.650 |
0.334 |
| 325 |
258468_at |
AT3G06070
|
unknown |
-0.648 |
0.158 |
| 326 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.646 |
0.284 |
| 327 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.646 |
0.333 |
| 328 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.638 |
-0.139 |
| 329 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.636 |
0.710 |
| 330 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.635 |
0.302 |
| 331 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.634 |
0.152 |
| 332 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
-0.633 |
-0.087 |
| 333 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.628 |
0.223 |
| 334 |
264899_at |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.623 |
0.019 |
| 335 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.622 |
0.124 |
| 336 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.620 |
0.333 |
| 337 |
264238_at |
AT1G54740
|
unknown |
-0.618 |
0.588 |
| 338 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.603 |
0.017 |
| 339 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.599 |
0.132 |
| 340 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.595 |
-0.010 |
| 341 |
258049_at |
AT3G16220
|
[Predicted eukaryotic LigT] |
-0.594 |
0.181 |
| 342 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.594 |
0.142 |
| 343 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.592 |
0.343 |
| 344 |
247326_at |
AT5G64110
|
[Peroxidase superfamily protein] |
-0.588 |
0.050 |
| 345 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.587 |
0.150 |
| 346 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.586 |
0.183 |
| 347 |
247754_at |
AT5G59080
|
unknown |
-0.583 |
-0.012 |
| 348 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.582 |
0.143 |
| 349 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.572 |
-0.187 |
| 350 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.572 |
-0.109 |
| 351 |
265892_at |
AT2G15020
|
unknown |
-0.570 |
0.049 |
| 352 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.568 |
0.229 |
| 353 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.567 |
0.213 |
| 354 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.554 |
-0.275 |
| 355 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.553 |
-0.046 |
| 356 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.550 |
0.137 |
| 357 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.549 |
0.004 |
| 358 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.543 |
0.173 |
| 359 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.542 |
0.039 |
| 360 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.541 |
-0.116 |
| 361 |
250327_at |
AT5G12050
|
unknown |
-0.539 |
0.154 |
| 362 |
258225_at |
AT3G15630
|
unknown |
-0.539 |
0.013 |
| 363 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.538 |
0.566 |
| 364 |
256548_at |
AT3G14770
|
AtSWEET2, SWEET2 |
-0.538 |
0.339 |
| 365 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.538 |
0.153 |
| 366 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.532 |
0.016 |
| 367 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.527 |
-0.016 |
| 368 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.524 |
0.183 |
| 369 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
-0.523 |
0.136 |
| 370 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.522 |
0.650 |
| 371 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.518 |
0.631 |
| 372 |
253425_at |
AT4G32190
|
[Myosin heavy chain-related protein] |
-0.514 |
-0.020 |
| 373 |
248624_at |
AT5G48790
|
unknown |
-0.511 |
0.096 |
| 374 |
246860_at |
AT5G25840
|
unknown |
-0.503 |
0.102 |
| 375 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.503 |
0.036 |
| 376 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.502 |
0.186 |
| 377 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.501 |
0.291 |
| 378 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.499 |
0.141 |
| 379 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.498 |
0.069 |
| 380 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
-0.493 |
0.147 |
| 381 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.492 |
0.049 |
| 382 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.491 |
0.745 |
| 383 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.488 |
0.147 |
| 384 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.482 |
0.003 |
| 385 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.479 |
-0.290 |
| 386 |
247474_at |
AT5G62280
|
unknown |
-0.478 |
0.166 |
| 387 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.477 |
0.212 |
| 388 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.477 |
0.250 |
| 389 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.476 |
0.097 |
| 390 |
253305_at |
AT4G33666
|
unknown |
-0.472 |
0.054 |
| 391 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.467 |
0.053 |
| 392 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.466 |
0.244 |
| 393 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.465 |
-0.012 |
| 394 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
-0.465 |
0.070 |
| 395 |
266984_at |
AT2G39570
|
ACR9, ACT domain repeats 9 |
-0.464 |
0.141 |
| 396 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.460 |
0.075 |
| 397 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.460 |
0.070 |
| 398 |
258275_at |
AT3G15760
|
unknown |
-0.459 |
-0.015 |
| 399 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.455 |
0.068 |
| 400 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.454 |
-0.007 |
| 401 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
-0.453 |
0.146 |
| 402 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
-0.453 |
0.007 |
| 403 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
-0.452 |
0.154 |
| 404 |
257315_at |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-0.451 |
0.317 |
| 405 |
263674_at |
AT2G04790
|
unknown |
-0.451 |
0.026 |
| 406 |
245349_at |
AT4G16690
|
ATMES16, ARABIDOPSIS THALIANA METHYL ESTERASE 16, MES16, methyl esterase 16 |
-0.450 |
0.239 |
| 407 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.449 |
-0.072 |
| 408 |
249932_at |
AT5G22390
|
unknown |
-0.444 |
0.020 |
| 409 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.443 |
-0.050 |
| 410 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.442 |
0.233 |
| 411 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.439 |
0.202 |
| 412 |
246069_at |
AT5G20220
|
[zinc knuckle (CCHC-type) family protein] |
-0.439 |
0.064 |
| 413 |
266717_at |
AT2G46735
|
unknown |
-0.438 |
0.100 |
| 414 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
-0.437 |
0.702 |
| 415 |
259373_at |
AT1G69160
|
unknown |
-0.436 |
0.233 |
| 416 |
262049_at |
AT1G80180
|
unknown |
-0.434 |
0.311 |
| 417 |
263118_at |
AT1G03090
|
MCCA |
-0.432 |
0.420 |
| 418 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.430 |
0.059 |
| 419 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.430 |
0.178 |
| 420 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.427 |
-0.015 |
| 421 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.426 |
0.901 |
| 422 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.425 |
0.309 |
| 423 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.425 |
0.358 |
| 424 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.422 |
0.018 |
| 425 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.422 |
0.161 |
| 426 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.421 |
0.591 |
| 427 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
-0.420 |
0.165 |
| 428 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.420 |
0.103 |
| 429 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.420 |
0.254 |
| 430 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.416 |
0.966 |
| 431 |
253125_at |
AT4G36040
|
DJC23, DNA J protein C23, J11, DnaJ11 |
-0.415 |
0.305 |
| 432 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.414 |
0.353 |
| 433 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.414 |
0.420 |
| 434 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.413 |
0.161 |
| 435 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.413 |
0.387 |
| 436 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.412 |
-0.228 |
| 437 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.411 |
0.352 |
| 438 |
249378_at |
AT5G40450
|
unknown |
-0.409 |
-0.252 |
| 439 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.409 |
-0.003 |
| 440 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.408 |
0.216 |
| 441 |
245011_at |
ATCG00430
|
PSBG, photosystem II reaction center protein G |
-0.408 |
-0.131 |
| 442 |
255626_at |
AT4G00780
|
[TRAF-like family protein] |
-0.408 |
0.127 |
| 443 |
260546_at |
AT2G43520
|
ATTI2, trypsin inhibitor protein 2, TI2, trypsin inhibitor protein 2 |
-0.408 |
0.990 |
| 444 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.407 |
0.060 |
| 445 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.405 |
0.088 |
| 446 |
255764_at |
AT1G16720
|
HCF173, high chlorophyll fluorescence phenotype 173 |
-0.405 |
0.102 |
| 447 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
-0.405 |
0.285 |
| 448 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
-0.404 |
0.187 |
| 449 |
264201_at |
AT1G22630
|
unknown |
-0.403 |
0.126 |
| 450 |
262194_at |
AT1G77930
|
DJC65, DNA J protein C65 |
-0.403 |
-0.001 |
| 451 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
-0.402 |
-0.173 |
| 452 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.401 |
0.081 |
| 453 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.401 |
0.023 |
| 454 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
-0.401 |
0.121 |
| 455 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.401 |
0.835 |
| 456 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.400 |
0.654 |
| 457 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.400 |
0.059 |
| 458 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.400 |
0.325 |
| 459 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.398 |
0.398 |
| 460 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.396 |
0.094 |
| 461 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.396 |
0.064 |
| 462 |
265611_at |
AT2G25510
|
unknown |
-0.396 |
0.154 |
| 463 |
252057_at |
AT3G52480
|
unknown |
-0.396 |
0.158 |
| 464 |
254954_at |
AT4G10910
|
unknown |
-0.395 |
0.338 |
| 465 |
251977_at |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-0.395 |
0.157 |
| 466 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.395 |
0.004 |
| 467 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.395 |
0.038 |
| 468 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.393 |
0.072 |
| 469 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.392 |
-0.011 |
| 470 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.391 |
0.220 |
| 471 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.391 |
0.662 |
| 472 |
254688_at |
AT4G13830
|
DJC26, DNA J protein C26, J20, DNAJ-like 20 |
-0.391 |
0.196 |
| 473 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.391 |
0.077 |
| 474 |
251036_at |
AT5G02160
|
unknown |
-0.390 |
0.068 |
| 475 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.388 |
1.291 |
| 476 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.388 |
-0.019 |
| 477 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.388 |
0.040 |
| 478 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.386 |
0.192 |
| 479 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.386 |
0.216 |
| 480 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.385 |
0.071 |
| 481 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
-0.384 |
0.175 |
| 482 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.384 |
0.263 |
| 483 |
265939_at |
AT2G19650
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.383 |
0.289 |
| 484 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.383 |
0.293 |
| 485 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.382 |
0.287 |
| 486 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.382 |
-0.307 |
| 487 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.381 |
0.150 |
| 488 |
251701_at |
AT3G56650
|
PPD6, PsbP-domain protein 6 |
-0.380 |
0.116 |
| 489 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.378 |
-0.127 |
| 490 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.377 |
0.162 |
| 491 |
257966_at |
AT3G19800
|
unknown |
-0.376 |
0.134 |
| 492 |
247931_at |
AT5G57040
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
-0.374 |
0.154 |
| 493 |
253243_at |
AT4G34560
|
unknown |
-0.374 |
0.102 |
| 494 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.373 |
0.123 |
| 495 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.373 |
0.113 |
| 496 |
250936_at |
AT5G03120
|
unknown |
-0.372 |
0.159 |
| 497 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.372 |
0.158 |
| 498 |
264725_at |
AT1G22885
|
unknown |
-0.372 |
0.109 |
| 499 |
246584_at |
AT5G14730
|
unknown |
-0.372 |
0.191 |
| 500 |
264738_at |
AT1G62250
|
unknown |
-0.371 |
-0.005 |
| 501 |
265481_at |
AT2G15960
|
unknown |
-0.370 |
1.191 |
| 502 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.370 |
0.210 |
| 503 |
256894_at |
AT3G21870
|
CYCP2;1, cyclin p2;1 |
-0.369 |
0.261 |
| 504 |
245010_at |
ATCG00420
|
NDHJ, NADH dehydrogenase subunit J |
-0.369 |
-0.151 |
| 505 |
244934_at |
ATCG01080
|
NDHG |
-0.368 |
-0.030 |
| 506 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.367 |
0.165 |
| 507 |
264774_at |
AT1G22890
|
unknown |
-0.366 |
-0.001 |
| 508 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-0.365 |
0.293 |
| 509 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.365 |
-0.000 |
| 510 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.364 |
0.095 |
| 511 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
-0.363 |
0.529 |
| 512 |
250223_at |
AT5G14070
|
ROXY2 |
-0.362 |
0.050 |
| 513 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
-0.362 |
0.192 |
| 514 |
264379_at |
AT2G25200
|
unknown |
-0.362 |
0.178 |
| 515 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.362 |
0.092 |
| 516 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.361 |
0.279 |
| 517 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.361 |
-0.135 |
| 518 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.360 |
-0.017 |
| 519 |
251142_at |
AT5G01015
|
unknown |
-0.360 |
-0.080 |
| 520 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.360 |
0.050 |
| 521 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.359 |
0.467 |
| 522 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
-0.359 |
0.139 |
| 523 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.358 |
-0.078 |
| 524 |
262456_at |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-0.358 |
-0.035 |
| 525 |
249029_at |
AT5G44870
|
LAZ5, LAZARUS 5, TTR1, tolerance to Tobacco ringspot virus 1 |
-0.358 |
-0.006 |
| 526 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.355 |
0.060 |
| 527 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.355 |
-0.078 |
| 528 |
250972_at |
AT5G02840
|
LCL1, LHY/CCA1-like 1 |
-0.353 |
0.243 |
| 529 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.353 |
0.543 |
| 530 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
-0.353 |
-0.010 |
| 531 |
267063_at |
AT2G41120
|
unknown |
-0.352 |
0.099 |
| 532 |
248756_at |
AT5G47560
|
ATSDAT, ATTDT, TONOPLAST DICARBOXYLATE TRANSPORTER, TDT, tonoplast dicarboxylate transporter |
-0.352 |
0.206 |
| 533 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
-0.352 |
0.103 |
| 534 |
248818_at |
AT5G47040
|
LON2, lon protease 2 |
-0.350 |
0.185 |
| 535 |
266402_at |
AT2G38780
|
unknown |
-0.350 |
-0.044 |
| 536 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.350 |
0.241 |
| 537 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-0.349 |
0.070 |
| 538 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-0.349 |
0.066 |
| 539 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.348 |
0.185 |
| 540 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.347 |
-0.196 |
| 541 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.346 |
0.091 |
| 542 |
262151_at |
AT1G52510
|
[alpha/beta-Hydrolases superfamily protein] |
-0.344 |
0.118 |
| 543 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.342 |
0.053 |
| 544 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.341 |
-0.056 |
| 545 |
263410_at |
AT2G04039
|
unknown |
-0.341 |
0.193 |
| 546 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.341 |
0.811 |
| 547 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
-0.341 |
-0.112 |
| 548 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.340 |
0.051 |
| 549 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.340 |
0.062 |
| 550 |
266188_at |
AT2G39000
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.340 |
0.217 |
| 551 |
250697_at |
AT5G06800
|
[myb-like HTH transcriptional regulator family protein] |
-0.340 |
-0.124 |
| 552 |
250217_at |
AT5G14120
|
[Major facilitator superfamily protein] |
-0.338 |
0.319 |
| 553 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.337 |
0.072 |
| 554 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.337 |
0.150 |
| 555 |
249941_at |
AT5G22270
|
unknown |
-0.336 |
-0.147 |
| 556 |
246028_at |
AT5G21170
|
5'-AMP-activated protein kinase beta-2 subunit protein |
-0.335 |
0.260 |
| 557 |
267057_at |
AT2G32500
|
[Stress responsive alpha-beta barrel domain protein] |
-0.335 |
0.001 |
| 558 |
247694_at |
AT5G59750
|
AtRIBA3, RIBA3, homolog of ribA 3 |
-0.335 |
0.090 |
| 559 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.334 |
0.248 |
| 560 |
267247_at |
AT2G30170
|
PBCP, PHOTOSYSTEM II CORE PHOSPHATASE |
-0.334 |
0.080 |
| 561 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.333 |
0.227 |
| 562 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.333 |
0.217 |
| 563 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.332 |
0.159 |
| 564 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.332 |
0.215 |
| 565 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.332 |
0.144 |
| 566 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.331 |
-0.021 |
| 567 |
253158_at |
AT4G35780
|
STY17, serine/threonine/tyrosine kinase 17 |
-0.330 |
0.163 |
| 568 |
260155_at |
AT1G52870
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
-0.330 |
0.115 |
| 569 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.330 |
0.093 |
| 570 |
257860_at |
AT3G13062
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.329 |
0.140 |
| 571 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.329 |
0.490 |
| 572 |
254924_at |
AT4G11330
|
ATMPK5, MAP kinase 5, MPK5, MAP kinase 5 |
-0.327 |
-0.411 |
| 573 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
-0.327 |
0.065 |
| 574 |
259275_at |
AT3G01060
|
unknown |
-0.327 |
0.106 |
| 575 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.326 |
0.713 |
| 576 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.326 |
0.130 |
| 577 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
-0.325 |
0.136 |
| 578 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
-0.323 |
0.168 |
| 579 |
262399_at |
AT1G49500
|
unknown |
-0.323 |
0.831 |
| 580 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
-0.323 |
0.289 |
| 581 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.322 |
0.192 |
| 582 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
-0.322 |
0.026 |
| 583 |
258375_at |
AT3G17470
|
ATCRSH, CRSH, Ca2+-activated RelA/spot homolog |
-0.321 |
0.083 |
| 584 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.321 |
0.115 |
| 585 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
-0.321 |
-0.114 |
| 586 |
263541_at |
AT2G24860
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
-0.320 |
0.131 |
| 587 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.320 |
-0.096 |
| 588 |
246158_at |
AT5G19855
|
AtRbcX2, RbcX2, homologue of cyanobacterial RbcX 2 |
-0.319 |
0.126 |
| 589 |
245906_at |
AT5G11070
|
unknown |
-0.318 |
0.168 |
| 590 |
267580_at |
AT2G41990
|
unknown |
-0.318 |
-0.014 |
| 591 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.318 |
-0.274 |
| 592 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
-0.317 |
0.125 |
| 593 |
262160_at |
AT1G52590
|
[Putative thiol-disulphide oxidoreductase DCC] |
-0.316 |
0.147 |
| 594 |
261143_at |
AT1G19770
|
ATPUP14, purine permease 14, PUP14, purine permease 14 |
-0.316 |
-0.046 |
| 595 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.315 |
0.137 |
| 596 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
-0.315 |
0.291 |
| 597 |
258939_at |
AT3G10020
|
unknown |
-0.315 |
0.100 |
| 598 |
245776_at |
AT1G30260
|
unknown |
-0.314 |
0.322 |
| 599 |
259015_at |
AT3G07350
|
unknown |
-0.313 |
0.578 |
| 600 |
245765_at |
AT1G33600
|
[Leucine-rich repeat (LRR) family protein] |
-0.312 |
-0.034 |