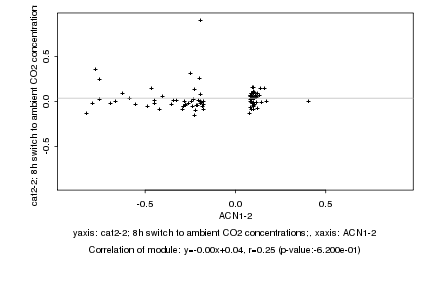
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ACN1-2 |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.400 |
0.007 |
| 2 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.172 |
0.009 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.158 |
0.152 |
| 4 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.139 |
-0.003 |
| 5 |
255836_at |
AT2G33440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.136 |
0.153 |
| 6 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.129 |
0.072 |
| 7 |
249225_at |
AT5G42140
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.120 |
0.094 |
| 8 |
247212_at |
AT5G65040
|
unknown |
0.118 |
-0.069 |
| 9 |
266410_at |
AT2G38770
|
EMB2765, EMBRYO DEFECTIVE 2765 |
0.115 |
0.053 |
| 10 |
259597_at |
AT1G27900
|
[RNA helicase family protein] |
0.114 |
0.060 |
| 11 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
0.113 |
-0.009 |
| 12 |
258043_at |
AT3G21290
|
[dentin sialophosphoprotein-related] |
0.109 |
0.073 |
| 13 |
259190_at |
AT3G01780
|
TPLATE |
0.102 |
0.037 |
| 14 |
245774_at |
AT1G30210
|
ATTCP24, TCP24, TEOSINTE BRANCHED 1, cycloidea, and PCF family 24 |
0.102 |
-0.028 |
| 15 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.101 |
0.104 |
| 16 |
245256_at |
AT4G15090
|
FAR1, FAR-RED IMPAIRED RESPONSE 1 |
0.100 |
0.094 |
| 17 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.100 |
0.101 |
| 18 |
263448_at |
AT2G31660
|
EMA1, enhanced miRNA activity 1, SAD2, SUPER SENSITIVE TO ABA AND DROUGHT2, URM9, UNARMED 9 |
0.099 |
-0.035 |
| 19 |
266431_at |
AT2G07230
|
[Mutator-like transposase family, has a 8.1e-59 P-value blast match to Q9SHN7 /450-633 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
0.099 |
-0.080 |
| 20 |
258566_at |
AT3G04110
|
ATGLR1.1, GLR1, GLUTAMATE RECEPTOR 1, GLR1.1, glutamate receptor 1.1 |
0.098 |
0.105 |
| 21 |
246233_at |
AT4G36550
|
[ARM repeat superfamily protein] |
0.098 |
0.000 |
| 22 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
0.096 |
0.038 |
| 23 |
256857_at |
AT3G15170
|
ANAC054, Arabidopsis NAC domain containing protein 54, ATNAC1, CUC1, CUP-SHAPED COTYLEDON1 |
0.096 |
-0.046 |
| 24 |
264858_at |
AT1G24190
|
ATSIN3, ARABIDOPSIS THALIANA SIN3 HOMOLOG, SIN3, ARABIDOPSIS THALIANA SIN3 HOMOLOG, SNL3, SIN3-like 3 |
0.096 |
0.165 |
| 25 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
0.095 |
0.117 |
| 26 |
264084_at |
AT2G31240
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.094 |
0.066 |
| 27 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
0.092 |
-0.020 |
| 28 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
0.091 |
0.162 |
| 29 |
253649_at |
AT4G29790
|
unknown |
0.090 |
0.062 |
| 30 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
0.089 |
-0.011 |
| 31 |
258844_at |
AT3G04740
|
ATMED14, ARABIDOPSIS MEDIATOR COMPONENTS 14, MED14, MEDIATOR COMPONENTS 14, SWP, STRUWWELPETER |
0.088 |
0.033 |
| 32 |
247659_at |
AT5G60040
|
NRPC1, nuclear RNA polymerase C1 |
0.088 |
0.035 |
| 33 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.087 |
0.099 |
| 34 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
0.086 |
0.023 |
| 35 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.086 |
0.061 |
| 36 |
262936_at |
AT1G79400
|
ATCHX2, cation/H+ exchanger 2, CHX2, cation/H+ exchanger 2 |
0.086 |
-0.009 |
| 37 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
0.085 |
0.091 |
| 38 |
265191_at |
AT1G05120
|
[Helicase protein with RING/U-box domain] |
0.085 |
0.075 |
| 39 |
246498_at |
AT5G16230
|
[Plant stearoyl-acyl-carrier-protein desaturase family protein] |
0.085 |
0.039 |
| 40 |
259175_at |
AT3G01560
|
unknown |
0.085 |
-0.080 |
| 41 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.083 |
0.070 |
| 42 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.080 |
-0.056 |
| 43 |
250711_at |
AT5G06110
|
[DnaJ domain ] |
0.080 |
0.069 |
| 44 |
263804_at |
AT2G40270
|
[Protein kinase family protein] |
0.078 |
0.056 |
| 45 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
0.078 |
0.028 |
| 46 |
247304_at |
AT5G63850
|
AAP4, amino acid permease 4 |
0.078 |
0.011 |
| 47 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.076 |
-0.127 |
| 48 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.825 |
-0.125 |
| 49 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.796 |
-0.020 |
| 50 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.779 |
0.357 |
| 51 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.756 |
0.248 |
| 52 |
247061_at |
AT5G66780
|
unknown |
-0.754 |
0.025 |
| 53 |
266544_at |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-0.694 |
-0.014 |
| 54 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
-0.669 |
0.005 |
| 55 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.631 |
0.092 |
| 56 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.591 |
0.044 |
| 57 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.554 |
-0.031 |
| 58 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
-0.490 |
-0.053 |
| 59 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.470 |
0.151 |
| 60 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.449 |
-0.021 |
| 61 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.449 |
0.021 |
| 62 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.421 |
-0.082 |
| 63 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.404 |
0.059 |
| 64 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.355 |
-0.025 |
| 65 |
247437_at |
AT5G62490
|
ATHVA22B, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE B, HVA22B, HVA22 homologue B |
-0.347 |
0.014 |
| 66 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.331 |
0.015 |
| 67 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
-0.295 |
-0.087 |
| 68 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
-0.290 |
-0.054 |
| 69 |
260859_at |
AT1G43780
|
scpl44, serine carboxypeptidase-like 44 |
-0.289 |
-0.047 |
| 70 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.286 |
0.009 |
| 71 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.280 |
-0.033 |
| 72 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
-0.278 |
-0.040 |
| 73 |
250624_at |
AT5G07330
|
unknown |
-0.260 |
-0.019 |
| 74 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.252 |
0.322 |
| 75 |
246242_at |
AT4G36600
|
[Late embryogenesis abundant (LEA) protein] |
-0.246 |
0.002 |
| 76 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
-0.238 |
-0.050 |
| 77 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.236 |
0.028 |
| 78 |
260668_at |
AT1G19530
|
unknown |
-0.232 |
-0.149 |
| 79 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
-0.227 |
0.134 |
| 80 |
266384_at |
AT2G14660
|
unknown |
-0.222 |
-0.090 |
| 81 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.219 |
-0.044 |
| 82 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.215 |
-0.040 |
| 83 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
-0.206 |
0.020 |
| 84 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-0.203 |
0.262 |
| 85 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.199 |
0.912 |
| 86 |
262373_at |
AT1G73120
|
unknown |
-0.199 |
-0.018 |
| 87 |
263881_at |
AT2G21820
|
unknown |
-0.196 |
0.006 |
| 88 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.196 |
0.081 |
| 89 |
250780_at |
AT5G05290
|
ATEXP2, ATEXPA2, expansin A2, ATHEXP ALPHA 1.12, EXP2, EXPANSIN 2, EXPA2, expansin A2 |
-0.191 |
-0.010 |
| 90 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.184 |
-0.051 |
| 91 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.178 |
-0.083 |
| 92 |
247456_at |
AT5G62160
|
AtZIP12, zinc transporter 12 precursor, ZIP12, zinc transporter 12 precursor |
-0.177 |
0.010 |
| 93 |
257880_at |
AT3G16910
|
AAE7, acyl-activating enzyme 7, ACN1, ACETATE NON-UTILIZING 1 |
-0.177 |
-0.029 |