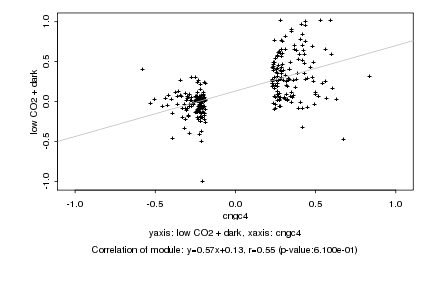
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cngc4 |
Log2 signal ratio
low CO2 + dark |
| 1 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.835 |
0.325 |
| 2 |
261221_at |
AT1G19960
|
unknown |
0.671 |
-0.468 |
| 3 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.627 |
0.026 |
| 4 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.602 |
0.165 |
| 5 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.595 |
0.592 |
| 6 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.589 |
1.018 |
| 7 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.563 |
0.044 |
| 8 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.559 |
0.258 |
| 9 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.558 |
0.663 |
| 10 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.537 |
0.233 |
| 11 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.526 |
1.027 |
| 12 |
246875_at |
AT5G26130
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.514 |
0.071 |
| 13 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
0.497 |
0.119 |
| 14 |
267262_at |
AT2G22990
|
SCPL8, SERINE CARBOXYPEPTIDASE-LIKE 8, SNG1, sinapoylglucose 1 |
0.496 |
0.089 |
| 15 |
257486_at |
AT1G63590
|
[Receptor-like protein kinase-related family protein] |
0.489 |
-0.029 |
| 16 |
247047_at |
AT5G66650
|
unknown |
0.485 |
0.254 |
| 17 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
0.481 |
0.499 |
| 18 |
259982_at |
AT1G76410
|
ATL8 |
0.479 |
0.697 |
| 19 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.478 |
0.309 |
| 20 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.463 |
0.435 |
| 21 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
0.446 |
0.298 |
| 22 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
0.445 |
-0.068 |
| 23 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.433 |
0.759 |
| 24 |
264238_at |
AT1G54740
|
unknown |
0.433 |
1.003 |
| 25 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
0.431 |
0.958 |
| 26 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.431 |
0.286 |
| 27 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.430 |
0.360 |
| 28 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.429 |
0.597 |
| 29 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.427 |
0.479 |
| 30 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
0.419 |
0.649 |
| 31 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.416 |
-0.077 |
| 32 |
259980_at |
AT1G76520
|
[Auxin efflux carrier family protein] |
0.415 |
-0.324 |
| 33 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
0.415 |
0.714 |
| 34 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.413 |
0.848 |
| 35 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
0.412 |
0.518 |
| 36 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.410 |
0.975 |
| 37 |
253352_at |
AT4G33710
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.404 |
-0.006 |
| 38 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.401 |
0.788 |
| 39 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.396 |
0.595 |
| 40 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
0.393 |
-0.083 |
| 41 |
245731_at |
AT1G73500
|
ATMKK9, MKK9, MAP kinase kinase 9 |
0.391 |
0.532 |
| 42 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
0.382 |
0.352 |
| 43 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.379 |
0.647 |
| 44 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.375 |
0.282 |
| 45 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.373 |
0.182 |
| 46 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
0.368 |
0.653 |
| 47 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.365 |
0.705 |
| 48 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.361 |
0.264 |
| 49 |
247052_at |
AT5G66700
|
ATHB53, ARABIDOPSIS THALIANA HOMEOBOX 53, HB-8, HOMEOBOX-8, HB53, homeobox 53 |
0.358 |
0.084 |
| 50 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.353 |
0.059 |
| 51 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.349 |
0.889 |
| 52 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
0.348 |
0.125 |
| 53 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.346 |
-0.004 |
| 54 |
262047_at |
AT1G80160
|
GLYI7, glyoxylase I 7 |
0.344 |
0.912 |
| 55 |
253322_at |
AT4G33980
|
unknown |
0.340 |
0.069 |
| 56 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.338 |
0.501 |
| 57 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
0.337 |
0.278 |
| 58 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.334 |
0.411 |
| 59 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.331 |
0.410 |
| 60 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.328 |
0.299 |
| 61 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.326 |
0.266 |
| 62 |
266712_at |
AT2G46750
|
AtGulLO2, GulLO2, L -gulono-1,4-lactone ( L -GulL) oxidase 2 |
0.324 |
0.014 |
| 63 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.323 |
0.037 |
| 64 |
265330_at |
AT2G18440
|
GUT15, GENE WITH UNSTABLE TRANSCRIPT 15 |
0.319 |
0.189 |
| 65 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.318 |
0.260 |
| 66 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
0.315 |
0.091 |
| 67 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
0.314 |
0.204 |
| 68 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.312 |
0.822 |
| 69 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
0.310 |
0.033 |
| 70 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
0.310 |
0.327 |
| 71 |
258939_at |
AT3G10020
|
unknown |
0.307 |
0.660 |
| 72 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.304 |
0.058 |
| 73 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
0.301 |
0.046 |
| 74 |
261558_at |
AT1G01770
|
unknown |
0.295 |
0.275 |
| 75 |
263490_at |
AT2G42620
|
MAX2, MORE AXILLARY BRANCHES 2, ORE9, ORESARA 9, PPS, PLEIOTROPIC PHOTOSIGNALING |
0.294 |
0.394 |
| 76 |
265387_at |
AT2G20670
|
unknown |
0.291 |
0.754 |
| 77 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.289 |
0.610 |
| 78 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.288 |
0.468 |
| 79 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
0.286 |
0.429 |
| 80 |
260668_at |
AT1G19530
|
unknown |
0.286 |
0.771 |
| 81 |
263118_at |
AT1G03090
|
MCCA |
0.284 |
0.657 |
| 82 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
0.284 |
0.385 |
| 83 |
258026_at |
AT3G19290
|
ABF4, ABRE binding factor 4, AREB2, ABA-RESPONSIVE ELEMENT BINDING PROTEIN 2 |
0.283 |
0.358 |
| 84 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.282 |
0.564 |
| 85 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.282 |
0.307 |
| 86 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
0.281 |
0.605 |
| 87 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.281 |
0.390 |
| 88 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
0.280 |
0.281 |
| 89 |
264580_at |
AT1G05340
|
unknown |
0.280 |
1.020 |
| 90 |
259316_at |
AT3G01175
|
unknown |
0.278 |
0.051 |
| 91 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
0.275 |
-0.050 |
| 92 |
259370_at |
AT1G69050
|
unknown |
0.274 |
0.110 |
| 93 |
264383_at |
AT2G25080
|
ATGPX1, GLUTATHIONE PEROXIDASE 1, GPX1, glutathione peroxidase 1 |
0.273 |
0.032 |
| 94 |
255893_at |
AT1G17960
|
[Threonyl-tRNA synthetase] |
0.270 |
0.025 |
| 95 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.270 |
-0.060 |
| 96 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.269 |
0.633 |
| 97 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.267 |
0.063 |
| 98 |
263157_at |
AT1G54100
|
ALDH7B4, aldehyde dehydrogenase 7B4 |
0.266 |
0.456 |
| 99 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.265 |
0.615 |
| 100 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.264 |
0.269 |
| 101 |
266583_at |
AT2G46220
|
[Uncharacterized conserved protein (DUF2358)] |
0.261 |
0.129 |
| 102 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
0.261 |
0.025 |
| 103 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
0.260 |
0.236 |
| 104 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.260 |
0.319 |
| 105 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.259 |
0.092 |
| 106 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.259 |
0.423 |
| 107 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.259 |
0.565 |
| 108 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
0.258 |
0.578 |
| 109 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
0.258 |
0.363 |
| 110 |
256455_at |
AT1G75190
|
unknown |
0.257 |
0.190 |
| 111 |
260875_at |
AT1G21410
|
SKP2A |
0.255 |
0.410 |
| 112 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.255 |
0.271 |
| 113 |
261250_at |
AT1G05890
|
ARI5, ARIADNE 5, ATARI5, ARABIDOPSIS ARIADNE 5 |
0.252 |
0.375 |
| 114 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.249 |
0.547 |
| 115 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.248 |
0.062 |
| 116 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
0.247 |
-0.087 |
| 117 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.247 |
-0.030 |
| 118 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.246 |
0.208 |
| 119 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.243 |
0.171 |
| 120 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
0.242 |
0.068 |
| 121 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.240 |
0.493 |
| 122 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.240 |
0.306 |
| 123 |
263475_at |
AT2G31945
|
unknown |
0.239 |
0.768 |
| 124 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
0.239 |
-0.093 |
| 125 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.237 |
0.494 |
| 126 |
267628_at |
AT2G42280
|
AKS3, ABA-responsive kinase substrate 3, FBH4, FLOWERING BHLH 4 |
0.236 |
0.203 |
| 127 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.236 |
0.390 |
| 128 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
0.235 |
-0.021 |
| 129 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.234 |
0.415 |
| 130 |
249824_at |
AT5G23380
|
unknown |
0.233 |
0.447 |
| 131 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.232 |
0.465 |
| 132 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
0.231 |
0.253 |
| 133 |
265495_at |
AT2G15695
|
unknown |
0.230 |
0.229 |
| 134 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
0.230 |
0.194 |
| 135 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.228 |
0.266 |
| 136 |
245543_at |
AT4G15260
|
[UDP-Glycosyltransferase superfamily protein] |
0.227 |
0.431 |
| 137 |
257866_at |
AT3G17770
|
[Dihydroxyacetone kinase] |
0.227 |
0.281 |
| 138 |
261821_at |
AT1G11530
|
ATCXXS1, C-terminal cysteine residue is changed to a serine 1, CXXS1, C-terminal cysteine residue is changed to a serine 1 |
0.226 |
0.235 |
| 139 |
266403_at |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.584 |
0.410 |
| 140 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
-0.530 |
-0.018 |
| 141 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
-0.510 |
0.036 |
| 142 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.457 |
-0.055 |
| 143 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
-0.438 |
0.038 |
| 144 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.427 |
-0.039 |
| 145 |
264640_at |
AT1G65680
|
ATEXPB2, expansin B2, ATHEXP BETA 1.4, EXPB2, expansin B2 |
-0.421 |
0.082 |
| 146 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.401 |
0.036 |
| 147 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.398 |
-0.460 |
| 148 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.395 |
-0.147 |
| 149 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.380 |
0.113 |
| 150 |
256926_at |
AT3G22540
|
unknown |
-0.366 |
-0.033 |
| 151 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.363 |
0.073 |
| 152 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.361 |
0.130 |
| 153 |
252232_at |
AT3G49760
|
AtbZIP5, basic leucine-zipper 5, bZIP5, basic leucine-zipper 5 |
-0.348 |
0.083 |
| 154 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-0.347 |
0.267 |
| 155 |
255608_at |
AT4G01140
|
unknown |
-0.341 |
0.067 |
| 156 |
253188_at |
AT4G35300
|
TMT2, tonoplast monosaccharide transporter2 |
-0.333 |
-0.083 |
| 157 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
-0.331 |
-0.193 |
| 158 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.323 |
-0.332 |
| 159 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.321 |
-0.028 |
| 160 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.314 |
-0.216 |
| 161 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.312 |
0.108 |
| 162 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.312 |
-0.081 |
| 163 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.310 |
-0.102 |
| 164 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
-0.309 |
0.023 |
| 165 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
-0.304 |
0.044 |
| 166 |
260112_at |
AT1G63310
|
unknown |
-0.303 |
-0.078 |
| 167 |
264842_at |
AT1G03700
|
[Uncharacterised protein family (UPF0497)] |
-0.302 |
0.067 |
| 168 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.299 |
0.056 |
| 169 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.298 |
-0.178 |
| 170 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
-0.293 |
-0.171 |
| 171 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.291 |
-0.057 |
| 172 |
258178_at |
AT3G21680
|
unknown |
-0.289 |
-0.025 |
| 173 |
264318_at |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.287 |
0.090 |
| 174 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.287 |
-0.392 |
| 175 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.281 |
0.018 |
| 176 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
-0.280 |
0.016 |
| 177 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.279 |
-0.062 |
| 178 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.279 |
0.311 |
| 179 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
-0.277 |
0.051 |
| 180 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.274 |
-0.018 |
| 181 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.264 |
-0.025 |
| 182 |
259774_at |
AT1G29520
|
[AWPM-19-like family protein] |
-0.260 |
-0.009 |
| 183 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.253 |
0.302 |
| 184 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.253 |
-0.134 |
| 185 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
-0.252 |
0.015 |
| 186 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.252 |
0.063 |
| 187 |
251944_at |
AT3G53510
|
ABCG20, ATP-binding cassette G20 |
-0.250 |
0.012 |
| 188 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.249 |
0.014 |
| 189 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.246 |
0.066 |
| 190 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.246 |
0.056 |
| 191 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.245 |
-0.125 |
| 192 |
247189_at |
AT5G65390
|
AGP7, arabinogalactan protein 7 |
-0.244 |
0.032 |
| 193 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
-0.244 |
-0.118 |
| 194 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.243 |
0.060 |
| 195 |
260643_at |
AT1G53270
|
ABCG10, ATP-binding cassette G10 |
-0.243 |
0.035 |
| 196 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.243 |
0.063 |
| 197 |
260332_at |
AT1G70470
|
unknown |
-0.242 |
-0.132 |
| 198 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.242 |
0.074 |
| 199 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.240 |
0.250 |
| 200 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.239 |
-0.129 |
| 201 |
260666_at |
AT1G19300
|
ATGATL1, GATL1, GALACTURONOSYLTRANSFERASE-LIKE 1, GLZ1, GAOLAOZHUANGREN 1, PARVUS, PARVUS |
-0.238 |
-0.099 |
| 202 |
263465_at |
AT2G31940
|
unknown |
-0.238 |
0.114 |
| 203 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.238 |
0.005 |
| 204 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.237 |
-0.198 |
| 205 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.235 |
-0.051 |
| 206 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.235 |
-0.081 |
| 207 |
260599_at |
AT1G55940
|
CYP708A1, cytochrome P450, family 708, subfamily A, polypeptide 1 |
-0.233 |
0.013 |
| 208 |
265083_at |
AT1G03820
|
unknown |
-0.233 |
-0.219 |
| 209 |
264937_at |
AT1G61130
|
SCPL32, serine carboxypeptidase-like 32 |
-0.232 |
0.047 |
| 210 |
252692_at |
AT3G43960
|
[Cysteine proteinases superfamily protein] |
-0.231 |
-0.100 |
| 211 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.231 |
0.270 |
| 212 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
-0.230 |
0.028 |
| 213 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
-0.229 |
-0.088 |
| 214 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
-0.229 |
-0.045 |
| 215 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.228 |
-0.088 |
| 216 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
-0.227 |
-0.003 |
| 217 |
246067_at |
AT5G19410
|
ABCG23, ATP-binding cassette G23 |
-0.226 |
0.072 |
| 218 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.226 |
-0.129 |
| 219 |
250949_at |
AT5G03510
|
[C2H2-type zinc finger family protein] |
-0.225 |
-0.025 |
| 220 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.225 |
0.010 |
| 221 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.225 |
-0.408 |
| 222 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
-0.224 |
0.036 |
| 223 |
257143_at |
AT3G20110
|
CYP705A20, cytochrome P450, family 705, subfamily A, polypeptide 20 |
-0.223 |
0.017 |
| 224 |
259596_at |
AT1G28130
|
GH3.17 |
-0.223 |
0.220 |
| 225 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
-0.222 |
-0.250 |
| 226 |
246535_at |
AT5G15900
|
TBL19, TRICHOME BIREFRINGENCE-LIKE 19 |
-0.221 |
-0.074 |
| 227 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
-0.220 |
0.010 |
| 228 |
263226_at |
AT1G30690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.220 |
0.022 |
| 229 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.219 |
0.062 |
| 230 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.219 |
0.067 |
| 231 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.219 |
-0.250 |
| 232 |
266362_at |
AT2G32430
|
[Galactosyltransferase family protein] |
-0.219 |
-0.131 |
| 233 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
-0.218 |
0.156 |
| 234 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.218 |
0.024 |
| 235 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
-0.218 |
-0.176 |
| 236 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.218 |
-0.190 |
| 237 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.217 |
0.023 |
| 238 |
257371_at |
AT2G47810
|
NF-YB5, nuclear factor Y, subunit B5 |
-0.216 |
-0.019 |
| 239 |
263282_at |
AT2G14095
|
unknown |
-0.216 |
-0.012 |
| 240 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.215 |
-0.015 |
| 241 |
252457_at |
AT3G47180
|
[RING/U-box superfamily protein] |
-0.215 |
0.011 |
| 242 |
254109_at |
AT4G25240
|
SKS1, SKU5 similar 1 |
-0.214 |
-0.199 |
| 243 |
249958_at |
AT5G18970
|
[AWPM-19-like family protein] |
-0.214 |
-0.016 |
| 244 |
263979_at |
AT2G42840
|
PDF1, protodermal factor 1 |
-0.214 |
-0.049 |
| 245 |
253165_at |
AT4G35320
|
unknown |
-0.214 |
-0.370 |
| 246 |
257824_at |
AT3G25290
|
[Auxin-responsive family protein] |
-0.213 |
-0.041 |
| 247 |
245629_at |
AT1G56580
|
SVB, SMALLER WITH VARIABLE BRANCHES |
-0.213 |
-0.149 |
| 248 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-0.213 |
0.030 |
| 249 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.212 |
-0.492 |
| 250 |
258560_at |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
-0.212 |
0.079 |
| 251 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.210 |
-0.077 |
| 252 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.210 |
-0.191 |
| 253 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.209 |
-0.999 |
| 254 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
-0.209 |
0.047 |
| 255 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
-0.207 |
-0.137 |
| 256 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
-0.205 |
0.023 |
| 257 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.205 |
-0.125 |
| 258 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.204 |
0.079 |
| 259 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.204 |
-0.050 |
| 260 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
-0.203 |
0.039 |
| 261 |
255528_at |
AT4G02090
|
unknown |
-0.203 |
0.094 |
| 262 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
-0.201 |
0.120 |
| 263 |
264891_at |
AT1G23200
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.199 |
0.242 |
| 264 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
-0.199 |
-0.052 |
| 265 |
247962_at |
AT5G56580
|
ANQ1, ARABIDOPSIS NQK1, ATMKK6, ARABIDOPSIS THALIANA MAP KINASE KINASE 6, MKK6, MAP kinase kinase 6 |
-0.198 |
-0.219 |
| 266 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.198 |
-0.167 |
| 267 |
250674_at |
AT5G07130
|
LAC13, laccase 13 |
-0.197 |
0.012 |
| 268 |
248370_at |
AT5G52170
|
HDG7, homeodomain GLABROUS 7 |
-0.197 |
0.057 |
| 269 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.195 |
-0.141 |
| 270 |
264909_at |
AT2G17300
|
unknown |
-0.193 |
-0.142 |
| 271 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
-0.192 |
-0.098 |
| 272 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
-0.192 |
0.015 |
| 273 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
-0.191 |
0.026 |
| 274 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.189 |
0.227 |
| 275 |
247268_at |
AT5G64080
|
AtXYP1, XYP1, xylogen protein 1 |
-0.187 |
-0.253 |