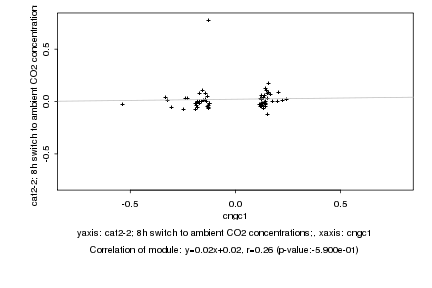
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
cngc1 |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
258585_at |
AT3G04340
|
emb2458, embryo defective 2458 |
0.240 |
0.026 |
| 2 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.223 |
0.010 |
| 3 |
256140_at |
AT1G48650
|
[DEA(D/H)-box RNA helicase family protein] |
0.201 |
0.090 |
| 4 |
261800_at |
AT1G30490
|
ATHB9, PHV, PHAVOLUTA |
0.198 |
0.001 |
| 5 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
0.173 |
0.002 |
| 6 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.165 |
0.074 |
| 7 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.157 |
0.177 |
| 8 |
263037_at |
AT1G23230
|
unknown |
0.157 |
0.088 |
| 9 |
259672_at |
AT1G68990
|
MGP3, male gametophyte defective 3 |
0.152 |
0.082 |
| 10 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
0.148 |
-0.122 |
| 11 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
0.148 |
0.035 |
| 12 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.146 |
0.113 |
| 13 |
255096_at |
AT4G08600
|
unknown |
0.141 |
-0.023 |
| 14 |
263324_at |
AT2G04220
|
unknown |
0.141 |
-0.045 |
| 15 |
252504_at |
AT3G46590
|
ATTRP2, TRFL1, TRF-like 1, TRP2 |
0.139 |
0.006 |
| 16 |
264126_at |
AT1G79280
|
AtTPR, TRANSLOCATED PROMOTER REGION, NUA, nuclear pore anchor |
0.139 |
0.127 |
| 17 |
258966_at |
AT3G10690
|
GYRA, DNA GYRASE A |
0.139 |
-0.012 |
| 18 |
264948_at |
AT1G77030
|
[hydrolases, acting on acid anhydrides, in phosphorus-containing anhydrides] |
0.136 |
-0.006 |
| 19 |
257196_at |
AT3G23790
|
AAE16, acyl activating enzyme 16 |
0.134 |
0.058 |
| 20 |
246257_at |
AT4G36690
|
ATU2AF65A |
0.133 |
0.044 |
| 21 |
252105_at |
AT3G51470
|
[Protein phosphatase 2C family protein] |
0.129 |
-0.061 |
| 22 |
261404_at |
AT1G18690
|
XXT4, xyloglucan xylosyltransferase 4 |
0.128 |
-0.011 |
| 23 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
0.126 |
-0.035 |
| 24 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.123 |
0.025 |
| 25 |
252182_at |
AT3G50670
|
U1-70K, U1 small nuclear ribonucleoprotein-70K, U1SNRNP |
0.123 |
0.023 |
| 26 |
252492_at |
AT3G46740
|
MAR1, MODIFIER OF ARG1 1, TOC75-III, translocon at the outer envelope membrane of chloroplasts 75-III |
0.122 |
-0.039 |
| 27 |
258766_at |
AT3G10700
|
GalAK, galacturonic acid kinase |
0.122 |
-0.014 |
| 28 |
264709_at |
AT1G09770
|
ATCDC5, ARABIDOPSIS THALIANA CELL DIVISION CYCLE 5, ATMYBCDC5, ARABIDOPSIS THALIANA MYB DOMAIN CELL DIVISION CYCLE 5, CDC5, cell division cycle 5 |
0.120 |
0.058 |
| 29 |
255382_at |
AT4G03430
|
EMB2770, EMBRYO DEFECTIVE 2770, STA1, STABILIZED 1 |
0.119 |
0.038 |
| 30 |
266033_at |
AT2G05830
|
MTI1, 5-METHYLTHIORIBOSE KINASE 1 |
0.119 |
-0.043 |
| 31 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.116 |
-0.024 |
| 32 |
250586_at |
AT5G07630
|
[lipid transporters] |
0.114 |
-0.020 |
| 33 |
264977_at |
AT1G27090
|
[glycine-rich protein] |
-0.541 |
-0.025 |
| 34 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
-0.337 |
0.045 |
| 35 |
253188_at |
AT4G35300
|
TMT2, tonoplast monosaccharide transporter2 |
-0.327 |
0.010 |
| 36 |
253830_at |
AT4G27652
|
unknown |
-0.308 |
-0.050 |
| 37 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.248 |
-0.069 |
| 38 |
245228_at |
AT3G29810
|
COBL2, COBRA-like protein 2 precursor |
-0.240 |
0.032 |
| 39 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
-0.230 |
0.033 |
| 40 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
-0.191 |
-0.011 |
| 41 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
-0.190 |
-0.070 |
| 42 |
251784_at |
AT3G55330
|
PPL1, PsbP-like protein 1 |
-0.188 |
-0.034 |
| 43 |
250431_at |
AT5G10440
|
CYCD4;2, cyclin d4;2 |
-0.185 |
-0.004 |
| 44 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.182 |
0.003 |
| 45 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.182 |
0.000 |
| 46 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.181 |
-0.057 |
| 47 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.174 |
0.081 |
| 48 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-0.174 |
-0.018 |
| 49 |
250949_at |
AT5G03510
|
[C2H2-type zinc finger family protein] |
-0.172 |
0.007 |
| 50 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
-0.162 |
0.004 |
| 51 |
254632_at |
AT4G18630
|
unknown |
-0.158 |
0.110 |
| 52 |
257371_at |
AT2G47810
|
NF-YB5, nuclear factor Y, subunit B5 |
-0.155 |
0.016 |
| 53 |
248250_at |
AT5G53130
|
ATCNGC1, CYCLIC NUCLEOTIDE-GATED CHANNEL 1, CNGC1, cyclic nucleotide gated channel 1 |
-0.145 |
0.025 |
| 54 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.143 |
0.079 |
| 55 |
247970_at |
AT5G56720
|
c-NAD-MDH3, cytosolic-NAD-dependent malate dehydrogenase 3 |
-0.138 |
0.005 |
| 56 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.136 |
-0.041 |
| 57 |
258631_at |
AT3G07970
|
QRT2, QUARTET 2 |
-0.135 |
0.052 |
| 58 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-0.132 |
0.783 |
| 59 |
259850_at |
AT1G72240
|
unknown |
-0.131 |
-0.063 |
| 60 |
264142_at |
AT1G78930
|
[Mitochondrial transcription termination factor family protein] |
-0.129 |
-0.035 |
| 61 |
246097_at |
AT5G20270
|
HHP1, heptahelical transmembrane protein1 |
-0.129 |
-0.048 |
| 62 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.128 |
-0.014 |
| 63 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.126 |
-0.017 |