|
probeID |
AGICode |
Annotation |
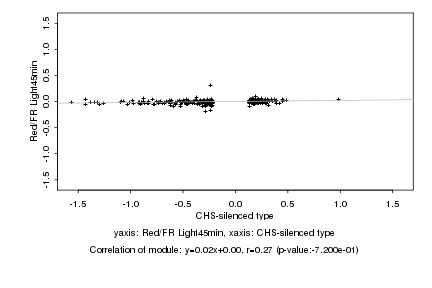
Log2 signal ratio
CHS-silenced type |
Log2 signal ratio
Red/FR Light45min |
| 1 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.975 |
0.040 |
| 2 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.485 |
0.028 |
| 3 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
0.452 |
0.022 |
| 4 |
264156_at |
AT1G65280
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.448 |
0.057 |
| 5 |
256021_at |
AT1G58270
|
ZW9 |
0.445 |
0.010 |
| 6 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.413 |
-0.023 |
| 7 |
267645_at |
AT2G32860
|
BGLU33, beta glucosidase 33 |
0.386 |
0.021 |
| 8 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.386 |
-0.034 |
| 9 |
259766_at |
AT1G64360
|
unknown |
0.381 |
0.005 |
| 10 |
266383_at |
AT2G14580
|
ATPRB1, basic pathogenesis-related protein 1, PRB1, basic pathogenesis-related protein 1 |
0.367 |
0.049 |
| 11 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.345 |
0.007 |
| 12 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.344 |
0.047 |
| 13 |
258434_at |
AT3G16770
|
ATEBP, ethylene-responsive element binding protein, EBP, ethylene-responsive element binding protein, ERF72, ETHYLENE RESPONSE FACTOR 72, RAP2.3, RELATED TO AP2 3 |
0.323 |
0.007 |
| 14 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.312 |
-0.061 |
| 15 |
250936_at |
AT5G03120
|
unknown |
0.306 |
0.021 |
| 16 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.305 |
0.044 |
| 17 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.300 |
0.037 |
| 18 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.298 |
0.004 |
| 19 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.288 |
-0.030 |
| 20 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.287 |
0.015 |
| 21 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
0.286 |
-0.022 |
| 22 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.280 |
0.051 |
| 23 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.277 |
0.038 |
| 24 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
0.268 |
0.006 |
| 25 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
0.260 |
-0.001 |
| 26 |
259611_at |
AT1G52280
|
AtRABG3d, RAB GTPase homolog G3D, RABG3d, RAB GTPase homolog G3D |
0.256 |
0.023 |
| 27 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.254 |
-0.019 |
| 28 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
0.253 |
-0.000 |
| 29 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
0.252 |
-0.014 |
| 30 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.250 |
-0.008 |
| 31 |
250881_at |
AT5G04080
|
unknown |
0.250 |
0.033 |
| 32 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
0.248 |
0.069 |
| 33 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.248 |
0.013 |
| 34 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
0.246 |
-0.002 |
| 35 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.240 |
-0.001 |
| 36 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
0.235 |
-0.023 |
| 37 |
249242_at |
AT5G42250
|
[Zinc-binding alcohol dehydrogenase family protein] |
0.233 |
0.007 |
| 38 |
263632_at |
AT2G04795
|
unknown |
0.232 |
-0.026 |
| 39 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
0.228 |
0.005 |
| 40 |
254317_at |
AT4G22510
|
unknown |
0.227 |
0.032 |
| 41 |
256397_at |
AT3G06110
|
ATMKP2, ARABIDOPSIS MAPK PHOSPHATASE 2, DSPTP1B, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 1B, MKP2, MAPK phosphatase 2 |
0.226 |
0.003 |
| 42 |
245710_at |
AT5G04330
|
CYP84A4, CYTOCHROME P450 84A4 |
0.223 |
0.005 |
| 43 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.223 |
-0.014 |
| 44 |
247091_at |
AT5G66390
|
PRX72, PEROXIDASE 72 |
0.219 |
-0.025 |
| 45 |
264288_at |
AT1G62045
|
unknown |
0.214 |
0.043 |
| 46 |
266860_at |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
0.214 |
-0.023 |
| 47 |
262686_at |
AT1G75990
|
[PAM domain (PCI/PINT associated module) protein] |
0.212 |
0.022 |
| 48 |
263756_at |
AT2G21270
|
UFD1, ubiquitin fusion degradation 1 |
0.211 |
0.040 |
| 49 |
247783_at |
AT5G58800
|
[Quinone reductase family protein] |
0.208 |
-0.018 |
| 50 |
266808_at |
AT2G29995
|
unknown |
0.208 |
-0.007 |
| 51 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.208 |
-0.020 |
| 52 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.208 |
0.010 |
| 53 |
267036_at |
AT2G38465
|
unknown |
0.206 |
-0.012 |
| 54 |
247074_at |
AT5G66590
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.206 |
0.017 |
| 55 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.206 |
-0.018 |
| 56 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
0.205 |
0.043 |
| 57 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.197 |
-0.007 |
| 58 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
0.197 |
0.024 |
| 59 |
256033_at |
AT1G07250
|
UGT71C4, UDP-glucosyl transferase 71C4 |
0.194 |
0.010 |
| 60 |
257646_at |
AT3G25740
|
MAP1B, methionine aminopeptidase 1C, MAP1C, METHIONINE AMINOPEPTIDASE 1C |
0.192 |
-0.018 |
| 61 |
263673_at |
AT2G04800
|
unknown |
0.191 |
0.002 |
| 62 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.191 |
-0.003 |
| 63 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.190 |
-0.025 |
| 64 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.190 |
-0.019 |
| 65 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.190 |
-0.002 |
| 66 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
0.189 |
0.008 |
| 67 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
0.189 |
0.011 |
| 68 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
0.189 |
0.043 |
| 69 |
253191_at |
AT4G35350
|
XCP1, xylem cysteine peptidase 1 |
0.187 |
0.002 |
| 70 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
0.187 |
-0.007 |
| 71 |
264010_at |
AT2G21100
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.186 |
0.098 |
| 72 |
251601_at |
AT3G57800
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.185 |
0.001 |
| 73 |
265051_at |
AT1G52100
|
[Mannose-binding lectin superfamily protein] |
0.184 |
-0.026 |
| 74 |
255710_at |
AT4G00030
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.183 |
0.041 |
| 75 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.183 |
0.036 |
| 76 |
256422_at |
AT1G33520
|
MOS2, modifier of snc1, 2 |
0.182 |
0.025 |
| 77 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
0.181 |
0.002 |
| 78 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
0.181 |
-0.040 |
| 79 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.179 |
-0.002 |
| 80 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.178 |
0.009 |
| 81 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
0.178 |
0.025 |
| 82 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
0.177 |
0.015 |
| 83 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
0.175 |
0.027 |
| 84 |
253810_at |
AT4G28220
|
NDB1, NAD(P)H dehydrogenase B1 |
0.175 |
-0.011 |
| 85 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.174 |
0.002 |
| 86 |
249072_at |
AT5G44060
|
unknown |
0.173 |
0.027 |
| 87 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
0.172 |
-0.007 |
| 88 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
0.171 |
0.015 |
| 89 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.171 |
0.009 |
| 90 |
250043_at |
AT5G18430
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.170 |
-0.027 |
| 91 |
258663_at |
AT3G08670
|
unknown |
0.168 |
-0.019 |
| 92 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
0.168 |
-0.008 |
| 93 |
247282_at |
AT5G64240
|
AtMC3, metacaspase 3, AtMCP1a, metacaspase 1a, MC3, metacaspase 3, MCP1a, metacaspase 1a |
0.168 |
-0.045 |
| 94 |
251752_at |
AT3G55740
|
ATPROT2, PROLINE TRANSPORTER 2, PROT2, proline transporter 2 |
0.167 |
-0.009 |
| 95 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
0.167 |
0.049 |
| 96 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
0.166 |
0.045 |
| 97 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.166 |
0.004 |
| 98 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.166 |
0.025 |
| 99 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.165 |
0.016 |
| 100 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.164 |
0.034 |
| 101 |
266791_at |
AT2G02960
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.164 |
-0.008 |
| 102 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
0.164 |
-0.007 |
| 103 |
260867_at |
AT1G43790
|
TED6, tracheary element differentiation-related 6 |
0.163 |
-0.008 |
| 104 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
0.163 |
-0.010 |
| 105 |
247124_at |
AT5G66060
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.162 |
-0.008 |
| 106 |
249882_at |
AT5G22890
|
STOP2, sensitive to proton rhizotoxicity 2 |
0.161 |
-0.010 |
| 107 |
248969_at |
AT5G45310
|
unknown |
0.161 |
0.016 |
| 108 |
252412_at |
AT3G47295
|
unknown |
0.160 |
0.010 |
| 109 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.159 |
0.014 |
| 110 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.158 |
0.027 |
| 111 |
246378_at |
AT1G57620
|
CYB, CYTOPLASMIC BODIES |
0.158 |
-0.003 |
| 112 |
247166_at |
AT5G65840
|
[Thioredoxin superfamily protein] |
0.156 |
0.029 |
| 113 |
260598_at |
AT1G55930
|
[CBS domain-containing protein / transporter associated domain-containing protein] |
0.156 |
0.059 |
| 114 |
260153_at |
AT1G52760
|
LysoPL2, lysophospholipase 2 |
0.156 |
-0.014 |
| 115 |
264270_at |
AT1G60260
|
BGLU5, beta glucosidase 5 |
0.155 |
-0.021 |
| 116 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
0.154 |
-0.030 |
| 117 |
261951_at |
AT1G64490
|
[DEK, chromatin associated protein] |
0.153 |
0.024 |
| 118 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.153 |
0.020 |
| 119 |
252419_at |
AT3G47510
|
unknown |
0.153 |
0.016 |
| 120 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
0.152 |
-0.032 |
| 121 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.152 |
0.028 |
| 122 |
249107_at |
AT5G43680
|
unknown |
0.151 |
0.015 |
| 123 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.150 |
0.012 |
| 124 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
0.148 |
0.009 |
| 125 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
0.148 |
-0.003 |
| 126 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.146 |
-0.002 |
| 127 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
0.146 |
-0.019 |
| 128 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
0.146 |
0.025 |
| 129 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
0.145 |
0.033 |
| 130 |
262010_at |
AT1G35612
|
[expressed protein] |
0.145 |
0.044 |
| 131 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
0.144 |
0.002 |
| 132 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
0.144 |
0.002 |
| 133 |
265843_at |
AT2G35690
|
ACX5, acyl-CoA oxidase 5 |
0.144 |
0.028 |
| 134 |
258501_at |
AT3G06780
|
[glycine-rich protein] |
0.143 |
0.021 |
| 135 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.143 |
-0.001 |
| 136 |
265902_at |
AT2G25590
|
[Plant Tudor-like protein] |
0.142 |
0.003 |
| 137 |
248407_at |
AT5G51500
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.142 |
0.016 |
| 138 |
256990_at |
AT3G28590
|
unknown |
0.142 |
-0.009 |
| 139 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
0.142 |
0.004 |
| 140 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.141 |
0.055 |
| 141 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.141 |
0.005 |
| 142 |
246891_at |
AT5G25490
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
0.140 |
-0.002 |
| 143 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
0.140 |
-0.012 |
| 144 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.140 |
0.005 |
| 145 |
262499_at |
AT1G21770
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.139 |
0.038 |
| 146 |
261893_at |
AT1G80690
|
[PPPDE putative thiol peptidase family protein] |
0.139 |
-0.001 |
| 147 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
0.139 |
0.019 |
| 148 |
264928_at |
AT1G60710
|
ATB2 |
0.139 |
0.011 |
| 149 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.138 |
0.038 |
| 150 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
0.137 |
0.011 |
| 151 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
0.137 |
0.001 |
| 152 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.136 |
0.020 |
| 153 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
0.135 |
0.012 |
| 154 |
245324_at |
AT4G17260
|
[Lactate/malate dehydrogenase family protein] |
0.135 |
0.006 |
| 155 |
250728_at |
AT5G06440
|
unknown |
0.135 |
0.024 |
| 156 |
256848_at |
AT3G27960
|
KLCR2, kinesin light chain-related 2 |
0.134 |
-0.018 |
| 157 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
0.134 |
-0.091 |
| 158 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
0.133 |
-0.032 |
| 159 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.133 |
0.003 |
| 160 |
262831_at |
AT1G14730
|
[Cytochrome b561/ferric reductase transmembrane protein family] |
0.133 |
0.035 |
| 161 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
0.133 |
-0.002 |
| 162 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.132 |
0.010 |
| 163 |
261112_at |
AT1G75420
|
[UDP-Glycosyltransferase superfamily protein] |
0.132 |
0.027 |
| 164 |
251645_at |
AT3G57790
|
[Pectin lyase-like superfamily protein] |
0.131 |
-0.003 |
| 165 |
264932_at |
AT1G61240
|
unknown |
0.130 |
0.024 |
| 166 |
260861_at |
AT1G43840
|
[CACTA-like transposase family (Ptta/En/Spm), has a 5.2e-121 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.571 |
-0.005 |
| 167 |
261186_at |
AT1G34530
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.8e-116 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.440 |
0.040 |
| 168 |
263675_x_at |
AT2G04770
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.0e-20 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.436 |
-0.052 |
| 169 |
255246_at |
AT4G05640
|
unknown |
-1.383 |
-0.016 |
| 170 |
265754_x_at |
AT2G10640
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.4e-42 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.345 |
-0.012 |
| 171 |
265600_at |
AT2G14230
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.0e-119 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.320 |
-0.002 |
| 172 |
261619_x_at |
AT1G33130
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-125 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.304 |
-0.044 |
| 173 |
266151_x_at |
AT2G12300
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.8e-41 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-1.265 |
-0.022 |
| 174 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-1.098 |
-0.014 |
| 175 |
263479_x_at |
AT2G04000
|
unknown |
-1.095 |
0.016 |
| 176 |
248949_at |
AT5G45570
|
[Ulp1 protease family protein] |
-1.077 |
0.007 |
| 177 |
253707_at |
AT4G29200
|
[Beta-galactosidase related protein] |
-1.033 |
-0.056 |
| 178 |
263338_at |
AT2G05000
|
unknown |
-1.018 |
-0.001 |
| 179 |
255918_at |
AT5G28570
|
unknown |
-0.984 |
0.020 |
| 180 |
259452_at |
AT1G44060
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.0e-22 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.977 |
-0.021 |
| 181 |
256748_x_at |
AT3G30396
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.1e-39 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.929 |
-0.011 |
| 182 |
250610_at |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-0.924 |
-0.016 |
| 183 |
262020_at |
AT1G35647
|
[gypsy-like retrotransposon family, has a 3.6e-209 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-0.924 |
-0.024 |
| 184 |
263346_at |
AT2G05660
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-0.918 |
-0.037 |
| 185 |
263819_x_at |
AT2G10140
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.7e-140 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
-0.909 |
-0.052 |
| 186 |
262031_x_at |
AT1G37160
|
[gypsy-like retrotransposon family (Athila), similar to putative Athila retroelement ORF1 protein GI:4567296 from (Arabidopsis thaliana)] |
-0.898 |
-0.015 |
| 187 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.886 |
0.059 |
| 188 |
246048_at |
AT5G28880
|
[pseudogene] |
-0.884 |
0.009 |
| 189 |
246828_at |
AT5G26350
|
unknown |
-0.877 |
-0.035 |
| 190 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
-0.847 |
-0.026 |
| 191 |
255172_x_at |
AT4G08000
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.0e-136 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
-0.837 |
0.010 |
| 192 |
249030_at |
AT5G44880
|
unknown |
-0.831 |
-0.025 |
| 193 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.798 |
0.048 |
| 194 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.787 |
-0.041 |
| 195 |
249259_at |
AT5G41660
|
unknown |
-0.775 |
-0.042 |
| 196 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.757 |
0.007 |
| 197 |
257057_at |
AT3G15310
|
unknown |
-0.736 |
-0.033 |
| 198 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.735 |
-0.017 |
| 199 |
256732_at |
AT3G30360
|
unknown |
-0.733 |
-0.014 |
| 200 |
263293_x_at |
AT2G10880
|
[hypothetical protein, similar to hypothetical protein GB:AAC26673] |
-0.722 |
0.016 |
| 201 |
265524_at |
AT2G06180
|
[pseudogene] |
-0.705 |
-0.019 |
| 202 |
249737_at |
AT5G24480
|
[Beta-galactosidase related protein] |
-0.683 |
-0.022 |
| 203 |
245032_at |
AT2G26630
|
[transposase IS4 family protein, contains Pfam profile: PF01609 transposase DDE domain] |
-0.679 |
-0.002 |
| 204 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.658 |
0.001 |
| 205 |
246810_at |
AT5G27160
|
unknown |
-0.646 |
-0.006 |
| 206 |
255235_at |
AT4G05510
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 4.0e-144 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.632 |
0.025 |
| 207 |
266375_at |
AT2G14630
|
unknown |
-0.628 |
0.017 |
| 208 |
250605_at |
AT5G07570
|
[glycine/proline-rich protein] |
-0.627 |
-0.026 |
| 209 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.621 |
-0.061 |
| 210 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.617 |
0.024 |
| 211 |
250979_at |
AT5G03090
|
unknown |
-0.616 |
-0.018 |
| 212 |
261235_x_at |
AT1G32840
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.594 |
-0.082 |
| 213 |
249114_at |
AT5G43800
|
[copia-like retrotransposon family, has a 0. P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
-0.580 |
-0.026 |
| 214 |
264075_at |
AT2G10850
|
unknown |
-0.579 |
-0.017 |
| 215 |
255343_at |
AT4G04530
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G27780.1)] |
-0.576 |
-0.041 |
| 216 |
261987_at |
AT1G33710
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.553 |
0.003 |
| 217 |
246698_at |
AT5G30480
|
[CACTA-like transposase family (Ptta/En/Spm), has a 6.2e-37 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.540 |
0.012 |
| 218 |
265365_at |
AT2G13310
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.3e-123 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.535 |
0.020 |
| 219 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.526 |
-0.078 |
| 220 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.525 |
-0.051 |
| 221 |
256449_at |
AT1G33460
|
[Mutator-like transposase family, has a 3.6e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.519 |
0.015 |
| 222 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.515 |
-0.010 |
| 223 |
258302_at |
AT3G30610
|
unknown |
-0.508 |
-0.008 |
| 224 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.507 |
-0.001 |
| 225 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.500 |
0.024 |
| 226 |
257777_x_at |
AT3G29210
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.495 |
-0.008 |
| 227 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-0.494 |
0.018 |
| 228 |
257284_at |
AT3G29650
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 5.1e-81 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
-0.491 |
0.003 |
| 229 |
259572_at |
AT1G20400
|
unknown |
-0.475 |
-0.028 |
| 230 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.474 |
0.004 |
| 231 |
246640_x_at |
AT5G34900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
-0.473 |
-0.054 |
| 232 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.470 |
-0.052 |
| 233 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.467 |
0.041 |
| 234 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.459 |
-0.047 |
| 235 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.455 |
-0.026 |
| 236 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.452 |
0.006 |
| 237 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.451 |
0.022 |
| 238 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
-0.448 |
-0.016 |
| 239 |
265503_at |
AT2G15510
|
[non-LTR retrotransposon family (LINE), has a 3.0e-20 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
-0.447 |
-0.003 |
| 240 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.436 |
-0.007 |
| 241 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.423 |
-0.013 |
| 242 |
249591_at |
AT5G37880
|
unknown |
-0.416 |
-0.032 |
| 243 |
252663_at |
AT3G44070
|
[Glycosyl hydrolase family 35 protein] |
-0.413 |
-0.008 |
| 244 |
251458_at |
AT3G60170
|
[copia-like retrotransposon family, has a 7.6e-229 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.410 |
-0.011 |
| 245 |
263851_at |
AT2G04460
|
unknown |
-0.407 |
0.004 |
| 246 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
-0.397 |
-0.037 |
| 247 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-0.396 |
0.004 |
| 248 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.394 |
-0.020 |
| 249 |
245834_at |
AT1G42200
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.3e-27 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.391 |
0.017 |
| 250 |
248727_at |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
-0.386 |
0.027 |
| 251 |
248728_at |
AT5G48000
|
CYP708 A2, CYTOCHROME P450, FAMILY 708, SUBFAMILY A, POLYPEPTIDE 2, CYP708A2, cytochrome P450, family 708, subfamily A, polypeptide 2, THAH, THALIANOL HYDROXYLASE, THAH1, THALIANOL HYDROXYLASE 1 |
-0.380 |
0.022 |
| 252 |
267273_at |
AT2G02520
|
[RNA-directed DNA polymerase (reverse transcriptase)-related family protein] |
-0.378 |
0.082 |
| 253 |
263089_at |
AT2G16000
|
[copia-like retrotransposon family, has a 2.6e-248 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.375 |
0.031 |
| 254 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-0.370 |
-0.042 |
| 255 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.365 |
0.007 |
| 256 |
252545_at |
AT3G45820
|
unknown |
-0.358 |
-0.024 |
| 257 |
263367_at |
AT2G20460
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.351 |
-0.041 |
| 258 |
252838_at |
AT3G42090
|
[contains domain LIN-9 RELATED (PTHR21689)] |
-0.348 |
0.011 |
| 259 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
-0.343 |
0.002 |
| 260 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.337 |
0.001 |
| 261 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.334 |
0.037 |
| 262 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
-0.330 |
-0.007 |
| 263 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.328 |
0.006 |
| 264 |
257354_x_at |
AT2G23480
|
unknown |
-0.321 |
0.015 |
| 265 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-0.319 |
-0.032 |
| 266 |
257111_x_at |
AT3G30450
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT3G30450.1)] |
-0.318 |
-0.021 |
| 267 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.318 |
0.011 |
| 268 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.316 |
-0.010 |
| 269 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.315 |
-0.094 |
| 270 |
259385_at |
AT1G13470
|
unknown |
-0.312 |
-0.018 |
| 271 |
258028_at |
AT3G27473
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.312 |
-0.010 |
| 272 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.311 |
0.035 |
| 273 |
245766_at |
AT1G33580
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 5.5e-96 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-0.306 |
-0.007 |
| 274 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.306 |
-0.008 |
| 275 |
266931_at |
AT2G07730
|
[non-LTR retrotransposon family (LINE), has a 5.0e-32 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.305 |
0.006 |
| 276 |
257566_x_at |
AT3G29610
|
unknown |
-0.300 |
0.024 |
| 277 |
265371_at |
AT2G06490
|
[CACTA-like transposase family (Ptta/En/Spm), has a 7.5e-83 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.299 |
0.018 |
| 278 |
262018_at |
AT1G35617
|
unknown |
-0.297 |
-0.070 |
| 279 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-0.294 |
-0.182 |
| 280 |
255932_at |
AT1G12720
|
[Mutator-like transposase family, has a 1.2e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.293 |
-0.034 |
| 281 |
252888_at |
AT4G39210
|
APL3 |
-0.292 |
-0.036 |
| 282 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-0.289 |
-0.028 |
| 283 |
250639_at |
AT5G07560
|
ATGRP20, GRP20, glycine-rich protein 20 |
-0.287 |
-0.078 |
| 284 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.282 |
0.006 |
| 285 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.280 |
0.004 |
| 286 |
258203_at |
AT3G13950
|
unknown |
-0.278 |
-0.064 |
| 287 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.275 |
-0.049 |
| 288 |
246869_at |
AT5G26020
|
[similar to Glutamic acid-rich protein precursor (GB:P13816)] |
-0.272 |
0.046 |
| 289 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.270 |
0.002 |
| 290 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.270 |
-0.008 |
| 291 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.267 |
0.012 |
| 292 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.267 |
-0.024 |
| 293 |
265752_at |
AT2G10660
|
[gypsy-like retrotransposon family (Athila), has a 4.3e-277 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.263 |
-0.009 |
| 294 |
257562_at |
AT3G30480
|
unknown |
-0.262 |
-0.030 |
| 295 |
252766_at |
AT3G42820
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G35770.1)] |
-0.260 |
-0.007 |
| 296 |
255917_at |
AT5G28560
|
unknown |
-0.259 |
-0.036 |
| 297 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.257 |
0.005 |
| 298 |
264995_at |
AT1G67240
|
[Mutator-like transposase family, has a 4.5e-23 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.253 |
-0.025 |
| 299 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
-0.252 |
0.019 |
| 300 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.251 |
0.021 |
| 301 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
-0.247 |
-0.029 |
| 302 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
-0.246 |
0.018 |
| 303 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.245 |
-0.056 |
| 304 |
245662_at |
AT1G28190
|
unknown |
-0.245 |
-0.010 |
| 305 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.244 |
-0.015 |
| 306 |
249726_at |
AT5G35480
|
unknown |
-0.244 |
0.015 |
| 307 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.241 |
0.310 |
| 308 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.241 |
0.019 |
| 309 |
259781_at |
AT1G29650
|
[non-LTR retrotransposon family (LINE), has a 1.5e-28 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
-0.241 |
-0.061 |
| 310 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
-0.240 |
-0.165 |
| 311 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.239 |
-0.051 |
| 312 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.239 |
-0.007 |
| 313 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.238 |
0.023 |
| 314 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.235 |
0.042 |
| 315 |
258957_at |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
-0.235 |
-0.023 |
| 316 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.234 |
0.026 |
| 317 |
263768_x_at |
AT2G06330
|
[pseudogene] |
-0.232 |
-0.008 |
| 318 |
255143_at |
AT4G08430
|
[Ulp1 protease family protein] |
-0.232 |
0.021 |
| 319 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.231 |
-0.093 |
| 320 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.231 |
0.002 |
| 321 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.230 |
-0.088 |
| 322 |
255414_at |
AT4G03156
|
[small GTPase-related] |
-0.230 |
-0.000 |
| 323 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.228 |
-0.064 |
| 324 |
255322_at |
AT4G04270
|
[CACTA-like transposase family (Ptta/En/Spm), has a 9.7e-78 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.227 |
-0.017 |
| 325 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
-0.227 |
-0.023 |
| 326 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.225 |
-0.018 |
| 327 |
263769_at |
AT2G06390
|
transposable element gene |
-0.224 |
-0.010 |
| 328 |
263511_at |
AT2G12420
|
[CACTA-like transposase family (Ptta/En/Spm), has a 3.9e-19 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.223 |
0.031 |
| 329 |
255463_at |
AT4G02960
|
ATRE2, retro element 2, RE2, retro element 2 |
-0.223 |
-0.017 |