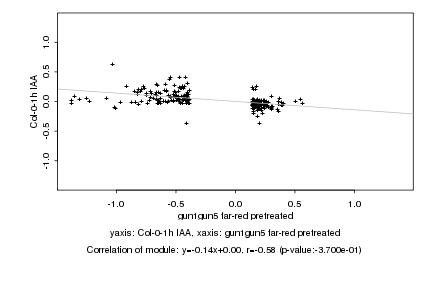
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gun1gun5 far-red pretreated |
Log2 signal ratio
Col-0-1h IAA |
| 1 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.563 |
-0.029 |
| 2 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.543 |
0.044 |
| 3 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.498 |
0.005 |
| 4 |
264822_at |
AT1G03457
|
AtBRN2, BRN2, Bruno-like 2 |
0.398 |
-0.029 |
| 5 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
0.391 |
-0.067 |
| 6 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
0.385 |
-0.064 |
| 7 |
255149_at |
AT4G08150
|
BP, BREVIPEDICELLUS, BP1, BREVIPEDICELLUS 1, KNAT1, KNOTTED-like from Arabidopsis thaliana |
0.379 |
-0.012 |
| 8 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.364 |
0.056 |
| 9 |
256815_at |
AT3G21380
|
[Mannose-binding lectin superfamily protein] |
0.361 |
-0.041 |
| 10 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
0.358 |
0.008 |
| 11 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.356 |
-0.160 |
| 12 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
0.348 |
-0.121 |
| 13 |
246926_at |
AT5G25240
|
unknown |
0.307 |
-0.073 |
| 14 |
255866_at |
AT2G30350
|
[Excinuclease ABC, C subunit, N-terminal] |
0.303 |
-0.100 |
| 15 |
257502_at |
AT1G78110
|
unknown |
0.302 |
-0.081 |
| 16 |
247581_at |
AT5G61350
|
[Protein kinase superfamily protein] |
0.300 |
0.096 |
| 17 |
255016_at |
AT4G10120
|
ATSPS4F |
0.299 |
-0.125 |
| 18 |
263765_at |
AT2G21540
|
ATSFH3, SEC14-LIKE 3, SFH3, SEC14-like 3 |
0.297 |
-0.116 |
| 19 |
265880_at |
AT2G42300
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.274 |
-0.006 |
| 20 |
265249_at |
AT2G01940
|
ATIDD15, ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 15, SGR5, SHOOT GRAVITROPISM 5 |
0.270 |
-0.085 |
| 21 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
0.269 |
-0.053 |
| 22 |
254193_at |
AT4G23870
|
unknown |
0.257 |
-0.074 |
| 23 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.256 |
0.007 |
| 24 |
262313_at |
AT1G70900
|
unknown |
0.249 |
-0.027 |
| 25 |
261626_at |
AT1G01990
|
unknown |
0.245 |
-0.044 |
| 26 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
0.243 |
-0.102 |
| 27 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.243 |
0.018 |
| 28 |
261941_at |
AT1G22490
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.241 |
0.014 |
| 29 |
248969_at |
AT5G45310
|
unknown |
0.241 |
-0.052 |
| 30 |
264821_at |
AT1G03470
|
NET3A, Networked 3A |
0.236 |
-0.034 |
| 31 |
248425_at |
AT5G51690
|
ACS12, 1-amino-cyclopropane-1-carboxylate synthase 12 |
0.233 |
0.029 |
| 32 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.232 |
-0.039 |
| 33 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.231 |
-0.030 |
| 34 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.228 |
-0.085 |
| 35 |
246244_at |
AT4G37250
|
[Leucine-rich repeat protein kinase family protein] |
0.222 |
-0.199 |
| 36 |
259984_at |
AT1G76460
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.219 |
-0.052 |
| 37 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
0.219 |
0.031 |
| 38 |
254142_at |
AT4G24630
|
[DHHC-type zinc finger family protein] |
0.217 |
-0.038 |
| 39 |
256413_at |
AT3G11100
|
[sequence-specific DNA binding transcription factors] |
0.214 |
-0.046 |
| 40 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.212 |
-0.135 |
| 41 |
256105_at |
AT1G16910
|
LSH8, LIGHT SENSITIVE HYPOCOTYLS 8 |
0.212 |
-0.110 |
| 42 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
0.211 |
-0.029 |
| 43 |
250548_at |
AT5G08120
|
MPB2C, movement protein binding protein 2C |
0.208 |
-0.035 |
| 44 |
258976_at |
AT3G01980
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.205 |
-0.044 |
| 45 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
0.203 |
0.006 |
| 46 |
259541_at |
AT1G20650
|
ASG5, ALTERED SEED GERMINATION 5 |
0.202 |
-0.120 |
| 47 |
248764_at |
AT5G47640
|
NF-YB2, nuclear factor Y, subunit B2 |
0.202 |
0.010 |
| 48 |
254693_at |
AT4G17880
|
MYC4 |
0.202 |
-0.062 |
| 49 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.201 |
-0.131 |
| 50 |
246963_at |
AT5G24820
|
[Eukaryotic aspartyl protease family protein] |
0.200 |
0.009 |
| 51 |
251353_at |
AT3G61080
|
[Protein kinase superfamily protein] |
0.199 |
-0.080 |
| 52 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
0.198 |
-0.366 |
| 53 |
252230_at |
AT3G49810
|
[ARM repeat superfamily protein] |
0.198 |
0.018 |
| 54 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.195 |
-0.022 |
| 55 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.194 |
-0.023 |
| 56 |
246155_at |
AT5G20030
|
[Plant Tudor-like RNA-binding protein] |
0.190 |
-0.073 |
| 57 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.190 |
0.015 |
| 58 |
253141_at |
AT4G35440
|
ATCLC-E, CLC-E, chloride channel E, CLCE, CHLORIDE CHANNEL E |
0.189 |
-0.126 |
| 59 |
260279_at |
AT1G80420
|
ATXRCC1, XRCC1, homolog of X-ray repair cross complementing 1 |
0.189 |
-0.053 |
| 60 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
0.187 |
-0.094 |
| 61 |
267637_at |
AT2G42190
|
unknown |
0.185 |
-0.143 |
| 62 |
256960_at |
AT3G13510
|
unknown |
0.184 |
-0.098 |
| 63 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.181 |
-0.091 |
| 64 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
0.181 |
-0.249 |
| 65 |
251306_at |
AT3G61260
|
[Remorin family protein] |
0.179 |
-0.005 |
| 66 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.178 |
0.049 |
| 67 |
258156_at |
AT3G18050
|
unknown |
0.178 |
-0.050 |
| 68 |
253225_at |
AT4G35020
|
ARAC3, RAC-like 3, ATROP6, RAC3, RAC-like 3, RHO1PS, ROP6, RHO-RELATED PROTEIN FROM PLANTS 6 |
0.175 |
-0.037 |
| 69 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
0.174 |
0.029 |
| 70 |
249109_at |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
0.174 |
0.260 |
| 71 |
248826_at |
AT5G47080
|
CKB1, casein kinase II beta chain 1 |
0.173 |
0.006 |
| 72 |
251225_at |
AT3G62660
|
GATL7, galacturonosyltransferase-like 7 |
0.172 |
-0.015 |
| 73 |
255540_at |
AT4G01800
|
AGY1, Albino or Glassy Yellow 1, AtcpSecA, Arabidopsis thaliana chloroplast SecA, SECA1 |
0.172 |
-0.024 |
| 74 |
259181_at |
AT3G01690
|
[alpha/beta-Hydrolases superfamily protein] |
0.171 |
-0.009 |
| 75 |
263053_at |
AT2G13440
|
[glucose-inhibited division family A protein] |
0.169 |
-0.053 |
| 76 |
260823_at |
AT1G06770
|
DRIP1, DREB2A-interacting protein 1 |
0.167 |
-0.032 |
| 77 |
262552_at |
AT1G31350
|
KUF1, KAR-UP F-box 1 |
0.167 |
0.213 |
| 78 |
260498_at |
AT2G41710
|
[Integrase-type DNA-binding superfamily protein] |
0.166 |
-0.074 |
| 79 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
0.164 |
-0.050 |
| 80 |
248251_at |
AT5G53220
|
unknown |
0.164 |
-0.088 |
| 81 |
263066_at |
AT2G17540
|
unknown |
0.163 |
-0.028 |
| 82 |
263673_at |
AT2G04800
|
unknown |
0.163 |
-0.034 |
| 83 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.163 |
-0.074 |
| 84 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
0.160 |
-0.105 |
| 85 |
259818_at |
AT1G49890
|
QWRF2, QWRF domain containing 2 |
0.160 |
-0.040 |
| 86 |
257651_at |
AT3G16850
|
[Pectin lyase-like superfamily protein] |
0.160 |
-0.036 |
| 87 |
245084_at |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
0.159 |
0.018 |
| 88 |
262322_at |
AT1G27590
|
unknown |
0.159 |
0.019 |
| 89 |
262728_at |
AT1G75820
|
ATCLV1, CLV1, CLAVATA 1, FAS3, FASCIATA 3, FLO5, FLOWER DEVELOPMENT 5 |
0.157 |
-0.022 |
| 90 |
251413_at |
AT3G60320
|
unknown |
0.157 |
-0.028 |
| 91 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.157 |
-0.092 |
| 92 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
0.155 |
0.016 |
| 93 |
250585_at |
AT5G07620
|
[Protein kinase superfamily protein] |
0.154 |
0.024 |
| 94 |
259476_at |
AT1G19000
|
[Homeodomain-like superfamily protein] |
0.154 |
-0.077 |
| 95 |
262043_at |
AT1G80190
|
PSF1, partner of SLD five 1 |
0.153 |
-0.008 |
| 96 |
253126_at |
AT4G36050
|
[endonuclease/exonuclease/phosphatase family protein] |
0.152 |
-0.111 |
| 97 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
0.152 |
-0.031 |
| 98 |
244995_at |
ATCG00150
|
ATPI |
0.151 |
-0.159 |
| 99 |
266990_at |
AT2G39190
|
ATATH8 |
0.151 |
0.040 |
| 100 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
0.151 |
-0.192 |
| 101 |
264336_at |
AT1G70360
|
[F-box family protein] |
0.150 |
-0.062 |
| 102 |
248394_at |
AT5G52070
|
[Agenet domain-containing protein] |
0.149 |
-0.011 |
| 103 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
0.149 |
0.216 |
| 104 |
262355_at |
AT1G72820
|
Mitochondrial substrate carrier family protein |
0.148 |
-0.116 |
| 105 |
249020_at |
AT5G44800
|
CHR4, chromatin remodeling 4, PKR1, PICKLE RELATED 1 |
0.147 |
-0.036 |
| 106 |
253484_at |
AT4G31720
|
STG1, SALT TOLERANCE DURING GERMINATION 1, TAF10, TBP-ASSOCIATED FACTOR 10, TAFII15, TBP-associated factor II 15 |
0.147 |
-0.029 |
| 107 |
262434_at |
AT1G47670
|
[Transmembrane amino acid transporter family protein] |
0.147 |
0.011 |
| 108 |
246296_at |
AT3G56750
|
unknown |
0.147 |
-0.021 |
| 109 |
261715_at |
AT1G18485
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.147 |
-0.057 |
| 110 |
267220_at |
AT2G02500
|
ATMEPCT, ISPD, MCT, 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLTRANSFERASE |
0.147 |
-0.035 |
| 111 |
264175_at |
AT1G02050
|
LAP6, LESS ADHESIVE POLLEN 6, PSKA, polyketide synthase A |
0.146 |
-0.069 |
| 112 |
261425_at |
AT1G18880
|
AtNPF2.9, NPF2.9, NRT1/ PTR family 2.9, NRT1.9, nitrate transporter 1.9 |
0.144 |
-0.072 |
| 113 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
0.144 |
-0.076 |
| 114 |
249482_at |
AT5G38980
|
unknown |
0.143 |
-0.043 |
| 115 |
251550_at |
AT3G58800
|
unknown |
0.142 |
-0.010 |
| 116 |
256564_at |
AT3G29770
|
ATMES11, ARABIDOPSIS THALIANA METHYL ESTERASE 11, MES11, methyl esterase 11 |
0.140 |
-0.109 |
| 117 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
0.140 |
-0.066 |
| 118 |
246701_at |
AT5G28020
|
ATCYSD2, CYSTEINE SYNTHASE D2, CYSD2, cysteine synthase D2 |
0.140 |
0.005 |
| 119 |
255522_at |
AT4G02260
|
AT-RSH1, RELA-SPOT HOMOLOG 1, ATRSH1, RELA/SPOT HOMOLOG 1, RSH1, RELA/SPOT homolog 1 |
0.139 |
-0.006 |
| 120 |
254669_at |
AT4G18370
|
DEG5, degradation of periplasmic proteins 5, DEGP5, DEGP PROTEASE 5, HHOA, PROTEASE HHOA PRECUSOR |
0.139 |
-0.065 |
| 121 |
259074_at |
AT3G02130
|
CLI1, clv3 peptide insensitive 1, RPK2, receptor-like protein kinase 2, TOAD2, TOADSTOOL 2 |
0.137 |
0.037 |
| 122 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.137 |
0.239 |
| 123 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
-1.384 |
0.024 |
| 124 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-1.378 |
-0.017 |
| 125 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
-1.359 |
0.087 |
| 126 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
-1.315 |
0.050 |
| 127 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-1.253 |
0.055 |
| 128 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
-1.233 |
0.011 |
| 129 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
-1.085 |
0.065 |
| 130 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
-1.039 |
0.638 |
| 131 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-1.022 |
-0.092 |
| 132 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-1.011 |
-0.113 |
| 133 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
-0.968 |
-0.009 |
| 134 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
-0.916 |
0.257 |
| 135 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
-0.881 |
-0.006 |
| 136 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.853 |
0.185 |
| 137 |
255811_at |
AT4G10250
|
ATHSP22.0 |
-0.847 |
-0.009 |
| 138 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.840 |
-0.003 |
| 139 |
259479_at |
AT1G19020
|
unknown |
-0.828 |
0.169 |
| 140 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.827 |
0.121 |
| 141 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.823 |
0.205 |
| 142 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-0.818 |
-0.044 |
| 143 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.804 |
0.173 |
| 144 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.792 |
0.011 |
| 145 |
246018_at |
AT5G10695
|
unknown |
-0.790 |
0.216 |
| 146 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
-0.778 |
0.262 |
| 147 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.771 |
0.222 |
| 148 |
254062_at |
AT4G25380
|
AtSAP10, Arabidopsis thaliana stress-associated protein 10, SAP10, stress-associated protein 10 |
-0.754 |
0.108 |
| 149 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.750 |
0.148 |
| 150 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.745 |
-0.024 |
| 151 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
-0.723 |
0.021 |
| 152 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.719 |
0.172 |
| 153 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.715 |
0.078 |
| 154 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
-0.709 |
0.145 |
| 155 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.696 |
0.066 |
| 156 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.676 |
0.122 |
| 157 |
247431_at |
AT5G62520
|
SRO5, similar to RCD one 5 |
-0.669 |
0.114 |
| 158 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.667 |
0.291 |
| 159 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.665 |
0.035 |
| 160 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.665 |
0.162 |
| 161 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
-0.662 |
-0.023 |
| 162 |
262085_at |
AT1G56060
|
unknown |
-0.660 |
0.274 |
| 163 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
-0.659 |
0.018 |
| 164 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
-0.654 |
0.033 |
| 165 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.652 |
0.166 |
| 166 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.651 |
-0.030 |
| 167 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.639 |
0.010 |
| 168 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.635 |
0.136 |
| 169 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.632 |
-0.031 |
| 170 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.630 |
0.057 |
| 171 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.611 |
0.011 |
| 172 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
-0.608 |
0.014 |
| 173 |
260522_x_at |
AT2G41730
|
unknown |
-0.604 |
0.187 |
| 174 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
-0.591 |
0.290 |
| 175 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.589 |
0.015 |
| 176 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.582 |
0.176 |
| 177 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
-0.581 |
0.198 |
| 178 |
260005_at |
AT1G67920
|
unknown |
-0.578 |
-0.004 |
| 179 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-0.568 |
0.005 |
| 180 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.565 |
0.117 |
| 181 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
-0.555 |
0.376 |
| 182 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
-0.553 |
0.084 |
| 183 |
257994_at |
AT3G19920
|
unknown |
-0.552 |
0.094 |
| 184 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
-0.547 |
0.415 |
| 185 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
-0.540 |
0.020 |
| 186 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.527 |
0.135 |
| 187 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
-0.527 |
0.024 |
| 188 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
-0.524 |
0.022 |
| 189 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.521 |
0.005 |
| 190 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
-0.517 |
0.274 |
| 191 |
252133_at |
AT3G50900
|
unknown |
-0.517 |
0.171 |
| 192 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.514 |
0.096 |
| 193 |
263515_at |
AT2G21640
|
unknown |
-0.511 |
0.073 |
| 194 |
264929_at |
AT1G60730
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.505 |
0.088 |
| 195 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
-0.505 |
0.153 |
| 196 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.498 |
0.034 |
| 197 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
-0.498 |
0.114 |
| 198 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
-0.483 |
0.040 |
| 199 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
-0.483 |
0.055 |
| 200 |
252278_at |
AT3G49530
|
ANAC062, NAC domain containing protein 62, NAC062, NAC domain containing protein 62, NTL6, NTM1 (NAC WITH TRANSMEMBRANE MOTIF 1)-LIKE 6 |
-0.482 |
0.172 |
| 201 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.481 |
0.100 |
| 202 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.478 |
0.050 |
| 203 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
-0.477 |
0.239 |
| 204 |
244950_at |
ATMG00160
|
COX2, cytochrome oxidase 2 |
-0.476 |
0.017 |
| 205 |
245209_at |
AT5G12340
|
unknown |
-0.474 |
0.410 |
| 206 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.468 |
0.236 |
| 207 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
-0.468 |
-0.009 |
| 208 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-0.463 |
-0.030 |
| 209 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.459 |
0.222 |
| 210 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
-0.459 |
0.097 |
| 211 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.453 |
-0.018 |
| 212 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-0.449 |
0.038 |
| 213 |
261430_at |
AT1G18830
|
SEC31A |
-0.447 |
-0.017 |
| 214 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-0.446 |
0.255 |
| 215 |
247177_at |
AT5G65300
|
unknown |
-0.446 |
0.259 |
| 216 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.439 |
0.227 |
| 217 |
247009_at |
AT5G67600
|
WIH1, WINDHOSE 1 |
-0.439 |
0.019 |
| 218 |
254190_at |
AT4G23885
|
unknown |
-0.438 |
0.034 |
| 219 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.437 |
0.092 |
| 220 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.436 |
0.093 |
| 221 |
246125_at |
AT5G19875
|
unknown |
-0.436 |
0.047 |
| 222 |
251259_at |
AT3G62260
|
[Protein phosphatase 2C family protein] |
-0.434 |
0.254 |
| 223 |
257226_at |
AT3G27880
|
unknown |
-0.431 |
0.108 |
| 224 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.426 |
0.041 |
| 225 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.422 |
0.410 |
| 226 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.422 |
0.026 |
| 227 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.419 |
0.029 |
| 228 |
247024_at |
AT5G66985
|
unknown |
-0.419 |
-0.360 |
| 229 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.415 |
-0.023 |
| 230 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
-0.412 |
0.120 |
| 231 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
-0.412 |
0.062 |
| 232 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.411 |
0.127 |
| 233 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
-0.410 |
0.086 |
| 234 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
-0.410 |
0.310 |
| 235 |
245243_at |
AT1G44414
|
unknown |
-0.406 |
-0.001 |
| 236 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.403 |
0.161 |
| 237 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.399 |
0.032 |
| 238 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.398 |
0.125 |
| 239 |
254263_at |
AT4G23493
|
unknown |
-0.395 |
-0.027 |
| 240 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
-0.395 |
0.045 |
| 241 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
-0.394 |
0.047 |
| 242 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
-0.393 |
0.201 |
| 243 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.392 |
-0.017 |