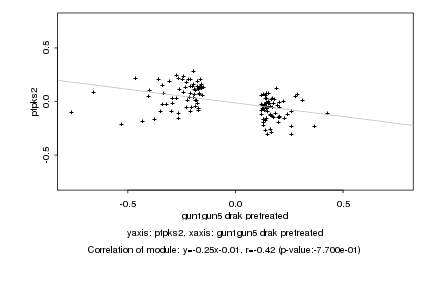
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gun1gun5 drak pretreated |
Log2 signal ratio
ptpks2 |
| 1 |
254945_at |
AT4G10940
|
[RING/U-box protein] |
0.427 |
-0.104 |
| 2 |
247324_at |
AT5G64190
|
unknown |
0.367 |
-0.225 |
| 3 |
253408_at |
AT4G32950
|
[Protein phosphatase 2C family protein] |
0.309 |
0.011 |
| 4 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
0.287 |
0.074 |
| 5 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
0.275 |
0.049 |
| 6 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.259 |
-0.307 |
| 7 |
263090_at |
AT2G16190
|
unknown |
0.256 |
-0.091 |
| 8 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.256 |
-0.224 |
| 9 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
0.238 |
-0.118 |
| 10 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.226 |
-0.157 |
| 11 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.221 |
0.004 |
| 12 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.204 |
-0.134 |
| 13 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.203 |
-0.050 |
| 14 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
0.201 |
0.000 |
| 15 |
245353_at |
AT4G16000
|
unknown |
0.201 |
-0.143 |
| 16 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.200 |
-0.191 |
| 17 |
251358_at |
AT3G61160
|
[Protein kinase superfamily protein] |
0.196 |
-0.140 |
| 18 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.191 |
-0.034 |
| 19 |
261626_at |
AT1G01990
|
unknown |
0.188 |
0.122 |
| 20 |
246725_at |
AT5G29040
|
[pseudogene] |
0.183 |
-0.104 |
| 21 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.177 |
0.019 |
| 22 |
265903_at |
AT2G25600
|
AKT6, SPIK, Shaker pollen inward K+ channel |
0.176 |
-0.141 |
| 23 |
265495_at |
AT2G15695
|
unknown |
0.173 |
-0.012 |
| 24 |
250482_at |
AT5G10320
|
unknown |
0.170 |
0.034 |
| 25 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
0.168 |
-0.050 |
| 26 |
256544_at |
AT1G42560
|
ATMLO9, ARABIDOPSIS THALIANA MILDEW RESISTANCE LOCUS O 9, MLO9, MILDEW RESISTANCE LOCUS O 9 |
0.168 |
-0.135 |
| 27 |
249729_at |
AT5G24410
|
PGL4, 6-phosphogluconolactonase 4 |
0.166 |
-0.281 |
| 28 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
0.166 |
0.026 |
| 29 |
246427_at |
AT5G17400
|
ER-ANT1, endoplasmic reticulum-adenine nucleotide transporter 1 |
0.162 |
-0.043 |
| 30 |
248015_at |
AT5G56370
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.161 |
-0.127 |
| 31 |
255545_at |
AT4G01890
|
[Pectin lyase-like superfamily protein] |
0.160 |
-0.114 |
| 32 |
265418_at |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
0.160 |
-0.261 |
| 33 |
253484_at |
AT4G31720
|
STG1, SALT TOLERANCE DURING GERMINATION 1, TAF10, TBP-ASSOCIATED FACTOR 10, TAFII15, TBP-associated factor II 15 |
0.155 |
-0.016 |
| 34 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
0.152 |
-0.018 |
| 35 |
264126_at |
AT1G79280
|
AtTPR, TRANSLOCATED PROMOTER REGION, NUA, nuclear pore anchor |
0.151 |
0.002 |
| 36 |
264096_at |
AT1G78995
|
unknown |
0.151 |
0.079 |
| 37 |
262199_at |
AT1G53800
|
unknown |
0.150 |
-0.053 |
| 38 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.148 |
-0.300 |
| 39 |
262549_at |
AT1G31290
|
AGO3, ARGONAUTE 3 |
0.146 |
-0.085 |
| 40 |
248776_at |
AT5G47900
|
unknown |
0.144 |
-0.017 |
| 41 |
263853_at |
AT2G04440
|
[MutT/nudix family protein] |
0.143 |
-0.048 |
| 42 |
249666_at |
AT5G35840
|
PHYC, phytochrome C |
0.142 |
0.032 |
| 43 |
266754_at |
AT2G46980
|
ASY3, ASYNAPTIC 3, AtASY3, Arabidopsis thaliana ASYNAPTIC 3 |
0.142 |
-0.064 |
| 44 |
257965_at |
AT3G19900
|
unknown |
0.141 |
-0.000 |
| 45 |
263255_at |
AT1G10490
|
unknown |
0.141 |
-0.046 |
| 46 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
0.141 |
0.076 |
| 47 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.140 |
-0.155 |
| 48 |
261773_at |
AT1G76250
|
unknown |
0.138 |
-0.045 |
| 49 |
267110_at |
AT2G14800
|
unknown |
0.138 |
-0.264 |
| 50 |
264301_at |
AT1G78780
|
[pathogenesis-related family protein] |
0.137 |
-0.174 |
| 51 |
259296_at |
AT3G05350
|
[Metallopeptidase M24 family protein] |
0.136 |
0.064 |
| 52 |
258565_at |
AT3G04350
|
unknown |
0.135 |
0.036 |
| 53 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.133 |
-0.082 |
| 54 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.131 |
-0.027 |
| 55 |
252109_at |
AT3G51540
|
unknown |
0.129 |
-0.223 |
| 56 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.129 |
-0.159 |
| 57 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.129 |
-0.063 |
| 58 |
259811_at |
AT1G49830
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.128 |
-0.162 |
| 59 |
256665_at |
AT3G20700
|
[F-box associated ubiquitination effector family protein] |
0.127 |
-0.187 |
| 60 |
245278_at |
AT4G17730
|
ATSYP23, SYP23, syntaxin of plants 23 |
0.127 |
0.066 |
| 61 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
0.124 |
-0.062 |
| 62 |
247531_at |
AT5G61550
|
[U-box domain-containing protein kinase family protein] |
0.123 |
-0.037 |
| 63 |
251540_at |
AT3G58740
|
CSY1, citrate synthase 1 |
0.121 |
-0.112 |
| 64 |
262974_at |
AT1G75550
|
[glycine-rich protein] |
0.120 |
-0.024 |
| 65 |
254653_at |
AT4G18180
|
[Pectin lyase-like superfamily protein] |
0.120 |
-0.082 |
| 66 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
0.120 |
0.057 |
| 67 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.765 |
-0.094 |
| 68 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.660 |
0.092 |
| 69 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
-0.531 |
-0.212 |
| 70 |
260028_at |
AT1G29980
|
unknown |
-0.466 |
0.220 |
| 71 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.432 |
-0.178 |
| 72 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.406 |
0.049 |
| 73 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.401 |
0.106 |
| 74 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.379 |
-0.162 |
| 75 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.358 |
0.212 |
| 76 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
-0.352 |
-0.091 |
| 77 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.340 |
-0.027 |
| 78 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.339 |
0.158 |
| 79 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.337 |
0.082 |
| 80 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
-0.321 |
-0.021 |
| 81 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.309 |
0.191 |
| 82 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.298 |
-0.088 |
| 83 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
-0.296 |
0.036 |
| 84 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.295 |
-0.012 |
| 85 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.278 |
0.037 |
| 86 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
-0.276 |
0.243 |
| 87 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.269 |
-0.108 |
| 88 |
259680_at |
AT1G77690
|
LAX3, like AUX1 3 |
-0.266 |
0.224 |
| 89 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.265 |
-0.155 |
| 90 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.260 |
0.115 |
| 91 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.250 |
0.209 |
| 92 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
-0.244 |
0.237 |
| 93 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
-0.244 |
0.087 |
| 94 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
-0.234 |
0.135 |
| 95 |
261129_at |
AT1G04820
|
TOR2, TORTIFOLIA 2, TUA4, tubulin alpha-4 chain |
-0.230 |
0.184 |
| 96 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
-0.228 |
-0.053 |
| 97 |
258576_at |
AT3G04230
|
[Ribosomal protein S5 domain 2-like superfamily protein] |
-0.226 |
0.012 |
| 98 |
246511_at |
AT5G15490
|
UGD3, UDP-glucose dehydrogenase 3 |
-0.221 |
0.212 |
| 99 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.217 |
0.045 |
| 100 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.212 |
-0.093 |
| 101 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.211 |
0.083 |
| 102 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
-0.210 |
0.149 |
| 103 |
260222_at |
AT1G74380
|
XXT5, xyloglucan xylosyltransferase 5 |
-0.209 |
0.208 |
| 104 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
-0.205 |
-0.053 |
| 105 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.202 |
0.148 |
| 106 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.197 |
0.166 |
| 107 |
265283_at |
AT2G20370
|
AtMUR3, KAM1, KATAMARI 1, MUR3, MURUS 3, RSA3, SHORT ROOT IN SALT MEDIUM 3 |
-0.197 |
0.284 |
| 108 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.195 |
0.045 |
| 109 |
251788_at |
AT3G55420
|
unknown |
-0.193 |
0.067 |
| 110 |
258040_at |
AT3G21190
|
AtMSR1, MSR1, Mannan Synthesis Related 1 |
-0.191 |
0.118 |
| 111 |
265411_at |
AT2G16630
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.187 |
0.111 |
| 112 |
257858_at |
AT3G12920
|
BRG3, BOI-related gene 3 |
-0.187 |
0.014 |
| 113 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
-0.186 |
-0.039 |
| 114 |
266297_at |
AT2G29570
|
ATPCNA2, A. THALIANA PROLIFERATING CELL NUCLEAR ANTIGEN 2, PCNA2, proliferating cell nuclear antigen 2 |
-0.182 |
0.017 |
| 115 |
251370_at |
AT3G60450
|
[Phosphoglycerate mutase family protein] |
-0.179 |
0.187 |
| 116 |
263002_at |
AT1G54200
|
unknown |
-0.178 |
0.146 |
| 117 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.178 |
-0.013 |
| 118 |
254030_at |
AT4G25890
|
[60S acidic ribosomal protein family] |
-0.176 |
0.115 |
| 119 |
262694_at |
AT1G62790
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.176 |
0.123 |
| 120 |
264007_at |
AT2G21140
|
ATPRP2, proline-rich protein 2, PRP2, proline-rich protein 2 |
-0.175 |
-0.063 |
| 121 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
-0.174 |
-0.077 |
| 122 |
250657_at |
AT5G07000
|
ATST2B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2B, ST2B, sulfotransferase 2B |
-0.169 |
0.076 |
| 123 |
260682_at |
AT1G17510
|
unknown |
-0.169 |
0.074 |
| 124 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
-0.167 |
0.135 |
| 125 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
-0.166 |
0.213 |
| 126 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.166 |
0.148 |
| 127 |
260590_at |
AT1G53310
|
ATPEPC1, ATPPC1, phosphoenolpyruvate carboxylase 1, PEPC1, PEP(PHOSPHOENOLPYRUVATE) CARBOXYLASE 1, PPC1, phosphoenolpyruvate carboxylase 1 |
-0.162 |
0.148 |
| 128 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
-0.161 |
0.126 |
| 129 |
267040_at |
AT2G34300
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.158 |
0.164 |
| 130 |
263809_at |
AT2G04570
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.154 |
0.062 |
| 131 |
266303_at |
AT2G27060
|
[Leucine-rich repeat protein kinase family protein] |
-0.152 |
0.132 |