|
probeID |
AGICode |
Annotation |
Log2 signal ratio
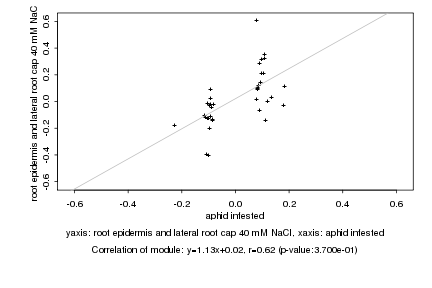
aphid infested |
Log2 signal ratio
root epidermis and lateral root cap 40 mM NaCl |
| 1 |
257288_at |
AT3G29670
|
PMAT2, phenolic glucoside malonyltransferase 2 |
0.182 |
0.114 |
| 2 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.177 |
-0.027 |
| 3 |
264636_at |
AT1G65490
|
unknown |
0.132 |
0.037 |
| 4 |
265336_at |
AT2G18290
|
APC10, anaphase promoting complex 10, AtAPC10, EMB2783, EMBRYO DEFECTIVE 2783 |
0.116 |
0.006 |
| 5 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
0.109 |
-0.139 |
| 6 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.108 |
0.326 |
| 7 |
248432_at |
AT5G51390
|
unknown |
0.107 |
0.356 |
| 8 |
262049_at |
AT1G80180
|
unknown |
0.102 |
0.212 |
| 9 |
257883_at |
AT3G16940
|
[calmodulin binding] |
0.097 |
0.319 |
| 10 |
267278_at |
AT2G19350
|
unknown |
0.097 |
0.211 |
| 11 |
259881_at |
AT1G76820
|
[eukaryotic translation initiation factor 2 (eIF-2) family protein] |
0.090 |
0.145 |
| 12 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.087 |
-0.060 |
| 13 |
248132_at |
AT5G54840
|
ATSGP1, SGP1 |
0.086 |
0.292 |
| 14 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.083 |
0.123 |
| 15 |
249739_at |
AT5G24520
|
ATTTG1, TRANSPARENT TESTA GLABRA 1, TTG, TTG1, TRANSPARENT TESTA GLABRA 1, URM23, UNARMED 23 |
0.080 |
0.092 |
| 16 |
247302_at |
AT5G63880
|
VPS20.1 |
0.080 |
0.103 |
| 17 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.079 |
0.112 |
| 18 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
0.078 |
0.614 |
| 19 |
251866_at |
AT3G54350
|
emb1967, embryo defective 1967 |
0.076 |
0.021 |
| 20 |
251899_at |
AT3G54400
|
[Eukaryotic aspartyl protease family protein] |
-0.228 |
-0.177 |
| 21 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
-0.118 |
-0.103 |
| 22 |
249006_at |
AT5G44660
|
unknown |
-0.113 |
-0.114 |
| 23 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.108 |
-0.396 |
| 24 |
266882_at |
AT2G44670
|
unknown |
-0.107 |
-0.010 |
| 25 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.106 |
-0.123 |
| 26 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
-0.103 |
-0.124 |
| 27 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.103 |
-0.398 |
| 28 |
266295_at |
AT2G29550
|
TUB7, tubulin beta-7 chain |
-0.098 |
-0.202 |
| 29 |
245384_at |
AT4G16790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.097 |
-0.027 |
| 30 |
264975_at |
AT1G27070
|
5'-AMP-activated protein kinase-related |
-0.095 |
0.029 |
| 31 |
247279_at |
AT5G64310
|
AGP1, arabinogalactan protein 1, ATAGP1 |
-0.095 |
0.097 |
| 32 |
265820_at |
AT2G17940
|
unknown |
-0.095 |
-0.018 |
| 33 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
-0.095 |
-0.110 |
| 34 |
255273_at |
AT4G05280
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G27780.1)] |
-0.092 |
-0.041 |
| 35 |
248948_at |
AT5G45560
|
[Pleckstrin homology (PH) domain-containing protein / lipid-binding START domain-containing protein] |
-0.089 |
-0.130 |
| 36 |
264228_at |
AT1G67490
|
GCS1, glucosidase 1, KNF, KNOPF |
-0.086 |
-0.141 |
| 37 |
255759_at |
AT1G16790
|
[ribosomal protein-related] |
-0.085 |
-0.022 |