|
probeID |
AGICode |
Annotation |
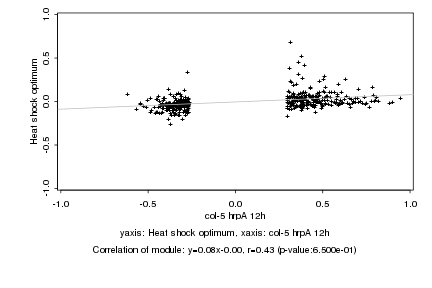
Log2 signal ratio
col-5 hrpA 12h |
Log2 signal ratio
Heat shock optimum |
| 1 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.943 |
0.037 |
| 2 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.897 |
0.000 |
| 3 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.880 |
-0.015 |
| 4 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.818 |
0.002 |
| 5 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.806 |
0.053 |
| 6 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.800 |
0.013 |
| 7 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.796 |
0.001 |
| 8 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.789 |
0.073 |
| 9 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.783 |
0.167 |
| 10 |
261394_at |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
0.776 |
0.002 |
| 11 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.763 |
-0.062 |
| 12 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.747 |
-0.012 |
| 13 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.743 |
-0.030 |
| 14 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.737 |
0.048 |
| 15 |
265705_at |
AT2G03410
|
[Mo25 family protein] |
0.722 |
-0.018 |
| 16 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.715 |
0.007 |
| 17 |
247913_at |
AT5G57510
|
unknown |
0.704 |
0.000 |
| 18 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.701 |
0.139 |
| 19 |
245209_at |
AT5G12340
|
unknown |
0.698 |
0.038 |
| 20 |
266779_at |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
0.686 |
-0.011 |
| 21 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.684 |
0.036 |
| 22 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.671 |
0.001 |
| 23 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.665 |
-0.020 |
| 24 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.659 |
0.024 |
| 25 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.656 |
-0.058 |
| 26 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.652 |
0.038 |
| 27 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.651 |
-0.024 |
| 28 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
0.640 |
-0.018 |
| 29 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.639 |
-0.013 |
| 30 |
254408_at |
AT4G21390
|
B120 |
0.635 |
0.060 |
| 31 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.625 |
0.264 |
| 32 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.623 |
0.033 |
| 33 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.611 |
-0.019 |
| 34 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.610 |
-0.002 |
| 35 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.607 |
-0.015 |
| 36 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.603 |
-0.015 |
| 37 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.603 |
0.115 |
| 38 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.599 |
0.021 |
| 39 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.595 |
-0.024 |
| 40 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.591 |
-0.016 |
| 41 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.587 |
-0.024 |
| 42 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.586 |
-0.038 |
| 43 |
263182_at |
AT1G05575
|
unknown |
0.586 |
0.207 |
| 44 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.581 |
0.068 |
| 45 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.580 |
0.090 |
| 46 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
0.578 |
0.024 |
| 47 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.577 |
-0.001 |
| 48 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.572 |
-0.020 |
| 49 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.563 |
-0.003 |
| 50 |
252470_at |
AT3G46930
|
[Protein kinase superfamily protein] |
0.562 |
0.105 |
| 51 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
0.550 |
0.003 |
| 52 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.549 |
-0.037 |
| 53 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.546 |
0.109 |
| 54 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.544 |
0.055 |
| 55 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.535 |
0.104 |
| 56 |
253796_at |
AT4G28460
|
unknown |
0.531 |
-0.034 |
| 57 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.527 |
0.021 |
| 58 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.523 |
0.067 |
| 59 |
245976_at |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
0.517 |
0.062 |
| 60 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.513 |
0.048 |
| 61 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.513 |
-0.018 |
| 62 |
254468_at |
AT4G20460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.512 |
-0.027 |
| 63 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.511 |
0.169 |
| 64 |
256499_at |
AT1G36640
|
unknown |
0.509 |
0.016 |
| 65 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.509 |
0.299 |
| 66 |
265186_at |
AT1G23560
|
unknown |
0.509 |
0.013 |
| 67 |
256177_at |
AT1G51620
|
[Protein kinase superfamily protein] |
0.506 |
0.036 |
| 68 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
0.505 |
0.110 |
| 69 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.499 |
0.125 |
| 70 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.499 |
0.258 |
| 71 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.498 |
0.044 |
| 72 |
247610_at |
AT5G60630
|
unknown |
0.497 |
-0.038 |
| 73 |
267226_at |
AT2G44010
|
unknown |
0.497 |
-0.055 |
| 74 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.493 |
0.015 |
| 75 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.489 |
-0.018 |
| 76 |
256799_at |
AT3G18560
|
unknown |
0.489 |
-0.075 |
| 77 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.487 |
-0.015 |
| 78 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.481 |
0.078 |
| 79 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.478 |
0.111 |
| 80 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.478 |
0.241 |
| 81 |
248060_at |
AT5G55560
|
[Protein kinase superfamily protein] |
0.476 |
0.001 |
| 82 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.474 |
0.036 |
| 83 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.471 |
0.063 |
| 84 |
255884_at |
AT1G20310
|
unknown |
0.471 |
-0.002 |
| 85 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.468 |
0.094 |
| 86 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.467 |
-0.005 |
| 87 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.465 |
0.029 |
| 88 |
247912_at |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.464 |
0.024 |
| 89 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.464 |
0.068 |
| 90 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.462 |
-0.006 |
| 91 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.461 |
0.047 |
| 92 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.458 |
-0.036 |
| 93 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.455 |
0.069 |
| 94 |
264773_at |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.454 |
-0.121 |
| 95 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.454 |
0.005 |
| 96 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.453 |
0.013 |
| 97 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.451 |
-0.062 |
| 98 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.447 |
-0.001 |
| 99 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.447 |
0.031 |
| 100 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
0.447 |
-0.033 |
| 101 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.444 |
-0.003 |
| 102 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.444 |
0.017 |
| 103 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
0.443 |
0.065 |
| 104 |
254571_at |
AT4G19370
|
unknown |
0.438 |
-0.052 |
| 105 |
251884_at |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.436 |
0.061 |
| 106 |
250267_at |
AT5G12930
|
unknown |
0.435 |
0.037 |
| 107 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.434 |
-0.035 |
| 108 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.429 |
0.059 |
| 109 |
264757_at |
AT1G61360
|
[S-locus lectin protein kinase family protein] |
0.428 |
-0.033 |
| 110 |
252484_at |
AT3G46690
|
[UDP-Glycosyltransferase superfamily protein] |
0.426 |
-0.025 |
| 111 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.426 |
0.164 |
| 112 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.425 |
0.154 |
| 113 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.425 |
0.049 |
| 114 |
248848_at |
AT5G46520
|
VICTR, VARIATION IN COMPOUND TRIGGERED ROOT growth response |
0.423 |
0.077 |
| 115 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
0.419 |
0.079 |
| 116 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.418 |
0.006 |
| 117 |
249252_at |
AT5G42010
|
[Transducin/WD40 repeat-like superfamily protein] |
0.418 |
-0.033 |
| 118 |
249057_at |
AT5G44480
|
DUR, DEFECTIVE UGE IN ROOT |
0.415 |
-0.034 |
| 119 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.415 |
0.074 |
| 120 |
247145_at |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
0.414 |
-0.010 |
| 121 |
260754_at |
AT1G49000
|
unknown |
0.410 |
-0.072 |
| 122 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.408 |
-0.010 |
| 123 |
258942_at |
AT3G09960
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
0.408 |
-0.022 |
| 124 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.406 |
0.018 |
| 125 |
253012_at |
AT4G37900
|
unknown |
0.404 |
-0.040 |
| 126 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.402 |
0.075 |
| 127 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.397 |
0.034 |
| 128 |
264660_at |
AT1G09940
|
HEMA2 |
0.397 |
0.048 |
| 129 |
258203_at |
AT3G13950
|
unknown |
0.396 |
0.113 |
| 130 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.395 |
0.034 |
| 131 |
245662_at |
AT1G28190
|
unknown |
0.395 |
0.032 |
| 132 |
251507_at |
AT3G59080
|
[Eukaryotic aspartyl protease family protein] |
0.394 |
0.075 |
| 133 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.392 |
0.418 |
| 134 |
252604_at |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
0.392 |
-0.035 |
| 135 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.389 |
-0.033 |
| 136 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
0.389 |
0.033 |
| 137 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.387 |
0.107 |
| 138 |
266658_at |
AT2G25735
|
unknown |
0.386 |
0.105 |
| 139 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.386 |
-0.008 |
| 140 |
252862_at |
AT4G39830
|
[Cupredoxin superfamily protein] |
0.384 |
0.037 |
| 141 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.384 |
0.097 |
| 142 |
253665_at |
AT4G30230
|
unknown |
0.383 |
-0.000 |
| 143 |
262762_at |
AT1G10700
|
PRS3, phosphoribosyl pyrophosphate (PRPP) synthase 3 |
0.383 |
-0.004 |
| 144 |
253124_at |
AT4G36030
|
ARO3, armadillo repeat only 3 |
0.383 |
-0.057 |
| 145 |
250018_at |
AT5G18150
|
[Methyltransferase-related protein] |
0.382 |
0.030 |
| 146 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.382 |
-0.064 |
| 147 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.381 |
-0.074 |
| 148 |
250323_at |
AT5G12880
|
[proline-rich family protein] |
0.380 |
-0.016 |
| 149 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.380 |
0.268 |
| 150 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.375 |
-0.099 |
| 151 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.373 |
-0.029 |
| 152 |
254605_at |
AT4G18950
|
[Integrin-linked protein kinase family] |
0.373 |
0.067 |
| 153 |
253654_at |
AT4G30060
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.373 |
-0.060 |
| 154 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.373 |
0.034 |
| 155 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.373 |
0.519 |
| 156 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
0.371 |
0.094 |
| 157 |
264766_at |
AT1G61420
|
[S-locus lectin protein kinase family protein] |
0.371 |
-0.035 |
| 158 |
262254_at |
AT1G53920
|
GLIP5, GDSL-motif lipase 5 |
0.371 |
-0.056 |
| 159 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.370 |
0.022 |
| 160 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
0.369 |
-0.061 |
| 161 |
255923_at |
AT1G22180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.368 |
0.010 |
| 162 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.367 |
0.021 |
| 163 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.364 |
0.056 |
| 164 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.364 |
0.026 |
| 165 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.361 |
0.322 |
| 166 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.360 |
0.457 |
| 167 |
252825_at |
AT4G39890
|
AtRABH1c, RAB GTPase homolog H1C, RABH1c, RAB GTPase homolog H1C |
0.359 |
0.090 |
| 168 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.357 |
0.035 |
| 169 |
257466_at |
AT1G62840
|
unknown |
0.355 |
0.053 |
| 170 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.352 |
0.006 |
| 171 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.350 |
-0.057 |
| 172 |
264774_at |
AT1G22890
|
unknown |
0.349 |
0.054 |
| 173 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
0.348 |
-0.037 |
| 174 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.348 |
0.199 |
| 175 |
266780_at |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
0.348 |
0.026 |
| 176 |
249767_at |
AT5G24090
|
ATCHIA, chitinase A, CHIA, chitinase A |
0.346 |
-0.047 |
| 177 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.342 |
0.017 |
| 178 |
248814_at |
AT5G46910
|
[Transcription factor jumonji (jmj) family protein / zinc finger (C5HC2 type) family protein] |
0.341 |
0.014 |
| 179 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.341 |
-0.059 |
| 180 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.339 |
0.002 |
| 181 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.337 |
-0.077 |
| 182 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.336 |
0.059 |
| 183 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.335 |
0.027 |
| 184 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.332 |
0.191 |
| 185 |
257478_at |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
0.330 |
0.097 |
| 186 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.329 |
0.023 |
| 187 |
251643_at |
AT3G57550
|
AGK2, guanylate kinase, GK-2, GUANYLATE KINAS 2 |
0.329 |
-0.036 |
| 188 |
256170_at |
AT1G51790
|
[Leucine-rich repeat protein kinase family protein] |
0.327 |
-0.016 |
| 189 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.327 |
0.085 |
| 190 |
264645_at |
AT1G08940
|
[Phosphoglycerate mutase family protein] |
0.325 |
0.052 |
| 191 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.323 |
-0.023 |
| 192 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.322 |
-0.007 |
| 193 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
0.321 |
-0.091 |
| 194 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.321 |
0.227 |
| 195 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.319 |
-0.034 |
| 196 |
254897_at |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
0.318 |
-0.044 |
| 197 |
249139_at |
AT5G43170
|
AZF3, zinc-finger protein 3, ZF3, zinc-finger protein 3 |
0.318 |
-0.007 |
| 198 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.317 |
0.065 |
| 199 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.315 |
-0.016 |
| 200 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.314 |
0.680 |
| 201 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.314 |
0.237 |
| 202 |
263108_at |
AT1G65240
|
[Eukaryotic aspartyl protease family protein] |
0.313 |
-0.012 |
| 203 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
0.312 |
-0.056 |
| 204 |
256442_at |
AT3G10930
|
unknown |
0.310 |
0.050 |
| 205 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.309 |
0.380 |
| 206 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
0.309 |
0.047 |
| 207 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.308 |
0.057 |
| 208 |
249779_at |
AT5G24230
|
[Lipase class 3-related protein] |
0.307 |
-0.087 |
| 209 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.306 |
0.104 |
| 210 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.305 |
0.035 |
| 211 |
246912_at |
AT5G25820
|
[Exostosin family protein] |
0.304 |
-0.003 |
| 212 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.302 |
0.120 |
| 213 |
254857_at |
AT4G12120
|
ATSEC1B, SEC1B |
0.302 |
-0.016 |
| 214 |
261545_at |
AT1G63530
|
unknown |
0.297 |
0.039 |
| 215 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.296 |
0.069 |
| 216 |
261501_at |
AT1G28390
|
[Protein kinase superfamily protein] |
0.296 |
0.015 |
| 217 |
249941_at |
AT5G22270
|
unknown |
0.294 |
-0.074 |
| 218 |
253178_at |
AT4G35170
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.294 |
-0.053 |
| 219 |
249532_at |
AT5G38780
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.294 |
-0.076 |
| 220 |
248306_at |
AT5G52830
|
ATWRKY27, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 27, WRKY27, WRKY DNA-binding protein 27 |
0.294 |
-0.170 |
| 221 |
263866_at |
AT2G36950
|
[Heavy metal transport/detoxification superfamily protein ] |
0.294 |
-0.006 |
| 222 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.621 |
0.082 |
| 223 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.568 |
-0.081 |
| 224 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.546 |
-0.015 |
| 225 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.545 |
-0.031 |
| 226 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.528 |
-0.051 |
| 227 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.511 |
-0.066 |
| 228 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
-0.504 |
0.017 |
| 229 |
262598_at |
AT1G15260
|
unknown |
-0.489 |
-0.121 |
| 230 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.487 |
0.046 |
| 231 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.486 |
-0.102 |
| 232 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.469 |
-0.068 |
| 233 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.458 |
-0.137 |
| 234 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.456 |
0.026 |
| 235 |
256603_at |
AT3G28270
|
unknown |
-0.453 |
-0.115 |
| 236 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.452 |
-0.121 |
| 237 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.451 |
0.033 |
| 238 |
253283_at |
AT4G34090
|
unknown |
-0.445 |
-0.059 |
| 239 |
261230_at |
AT1G20010
|
TUB5, tubulin beta-5 chain |
-0.443 |
0.068 |
| 240 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.440 |
0.003 |
| 241 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
-0.436 |
-0.135 |
| 242 |
250856_at |
AT5G04810
|
[pentatricopeptide (PPR) repeat-containing protein] |
-0.431 |
-0.050 |
| 243 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.430 |
-0.033 |
| 244 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.427 |
-0.040 |
| 245 |
263674_at |
AT2G04790
|
unknown |
-0.425 |
-0.133 |
| 246 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.422 |
-0.123 |
| 247 |
245348_at |
AT4G17770
|
ATTPS5, trehalose phosphatase/synthase 5, TPS5, TREHALOSE -6-PHOSPHATASE SYNTHASE S5, TPS5, trehalose phosphatase/synthase 5 |
-0.420 |
-0.073 |
| 248 |
245876_at |
AT1G26230
|
Cpn60beta4, chaperonin-60beta4 |
-0.419 |
-0.016 |
| 249 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.417 |
0.016 |
| 250 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.417 |
0.036 |
| 251 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.412 |
-0.038 |
| 252 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.412 |
-0.072 |
| 253 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.408 |
0.041 |
| 254 |
264672_at |
AT1G09750
|
[Eukaryotic aspartyl protease family protein] |
-0.407 |
0.040 |
| 255 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
-0.405 |
-0.023 |
| 256 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.403 |
-0.034 |
| 257 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.400 |
-0.020 |
| 258 |
252353_at |
AT3G48200
|
unknown |
-0.399 |
-0.098 |
| 259 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.399 |
-0.069 |
| 260 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.398 |
-0.026 |
| 261 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.396 |
-0.048 |
| 262 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.394 |
-0.034 |
| 263 |
255957_at |
AT1G22160
|
unknown |
-0.392 |
-0.097 |
| 264 |
257974_at |
AT3G20820
|
[Leucine-rich repeat (LRR) family protein] |
-0.392 |
-0.032 |
| 265 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.389 |
-0.079 |
| 266 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
-0.387 |
0.144 |
| 267 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.387 |
-0.068 |
| 268 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.386 |
-0.061 |
| 269 |
260347_at |
AT1G69420
|
[DHHC-type zinc finger family protein] |
-0.385 |
-0.032 |
| 270 |
253650_at |
AT4G30020
|
[PA-domain containing subtilase family protein] |
-0.385 |
-0.039 |
| 271 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.385 |
-0.203 |
| 272 |
262693_at |
AT1G62780
|
unknown |
-0.384 |
-0.019 |
| 273 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.383 |
-0.047 |
| 274 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.383 |
-0.020 |
| 275 |
253218_at |
AT4G34980
|
SLP2, subtilisin-like serine protease 2 |
-0.382 |
-0.018 |
| 276 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.380 |
-0.033 |
| 277 |
246231_at |
AT4G37080
|
unknown |
-0.380 |
-0.101 |
| 278 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.379 |
-0.019 |
| 279 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
-0.379 |
-0.056 |
| 280 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.377 |
-0.257 |
| 281 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
-0.376 |
0.091 |
| 282 |
257717_at |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.374 |
-0.023 |
| 283 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.374 |
-0.136 |
| 284 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
-0.372 |
-0.021 |
| 285 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.372 |
-0.068 |
| 286 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.371 |
-0.037 |
| 287 |
265892_at |
AT2G15020
|
unknown |
-0.371 |
-0.156 |
| 288 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.368 |
-0.037 |
| 289 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.366 |
-0.077 |
| 290 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.362 |
-0.096 |
| 291 |
261480_at |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.361 |
-0.039 |
| 292 |
253545_at |
AT4G31310
|
[AIG2-like (avirulence induced gene) family protein] |
-0.359 |
-0.080 |
| 293 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.358 |
-0.047 |
| 294 |
251193_at |
AT3G62910
|
APG3, ALBINO AND PALE GREEN, AtcpRF1, Arabidopsis thaliana chloroplast RF1, cpRF1, chloroplast ribosome release factor 1 |
-0.358 |
-0.013 |
| 295 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.358 |
0.064 |
| 296 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.356 |
-0.034 |
| 297 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
-0.356 |
-0.123 |
| 298 |
263597_at |
AT2G01870
|
unknown |
-0.354 |
-0.008 |
| 299 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
-0.353 |
-0.038 |
| 300 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.353 |
0.000 |
| 301 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
-0.352 |
-0.152 |
| 302 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.350 |
-0.091 |
| 303 |
246792_at |
AT5G27290
|
unknown |
-0.349 |
-0.036 |
| 304 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.348 |
-0.055 |
| 305 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.347 |
-0.138 |
| 306 |
252363_at |
AT3G48460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.346 |
-0.135 |
| 307 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.346 |
-0.046 |
| 308 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.345 |
0.042 |
| 309 |
257773_at |
AT3G29185
|
unknown |
-0.343 |
-0.013 |
| 310 |
254638_at |
AT4G18740
|
[Rho termination factor] |
-0.343 |
0.018 |
| 311 |
257533_at |
AT3G10840
|
[alpha/beta-Hydrolases superfamily protein] |
-0.342 |
-0.101 |
| 312 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.341 |
-0.026 |
| 313 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.341 |
-0.061 |
| 314 |
248095_at |
AT5G55230
|
ATMAP65-1, microtubule-associated proteins 65-1, MAP65-1, MAP65-1, microtubule-associated proteins 65-1 |
-0.340 |
0.015 |
| 315 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.339 |
-0.048 |
| 316 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.339 |
0.083 |
| 317 |
266608_at |
AT2G35500
|
SKL2, shikimate kinase-like 2 |
-0.336 |
-0.000 |
| 318 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
-0.336 |
-0.060 |
| 319 |
250739_at |
AT5G05740
|
ATEGY2, EGY2, ethylene-dependent gravitropism-deficient and yellow-green-like 2 |
-0.334 |
-0.009 |
| 320 |
267356_at |
AT2G39930
|
ATISA1, ARABIDOPSIS THALIANA ISOAMYLASE 1, ISA1, isoamylase 1 |
-0.333 |
-0.008 |
| 321 |
266575_at |
AT2G24060
|
[Translation initiation factor 3 protein] |
-0.333 |
-0.017 |
| 322 |
258800_at |
AT3G04550
|
unknown |
-0.332 |
-0.007 |
| 323 |
262397_at |
AT1G49380
|
[cytochrome c biogenesis protein family] |
-0.330 |
-0.075 |
| 324 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.329 |
-0.070 |
| 325 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.328 |
0.096 |
| 326 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
-0.328 |
-0.006 |
| 327 |
254427_at |
AT4G21190
|
emb1417, embryo defective 1417 |
-0.328 |
-0.015 |
| 328 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.328 |
-0.152 |
| 329 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.327 |
0.020 |
| 330 |
253165_at |
AT4G35320
|
unknown |
-0.326 |
-0.150 |
| 331 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
-0.325 |
-0.008 |
| 332 |
248402_at |
AT5G52100
|
CRR1, chlororespiration reduction 1 |
-0.325 |
-0.041 |
| 333 |
247439_at |
AT5G62670
|
AHA11, H(+)-ATPase 11, HA11, H(+)-ATPase 11 |
-0.324 |
-0.070 |
| 334 |
264930_at |
AT1G60800
|
AtNIK3, NIK3, NSP-interacting kinase 3 |
-0.324 |
-0.070 |
| 335 |
264096_at |
AT1G78995
|
unknown |
-0.323 |
-0.088 |
| 336 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.323 |
-0.004 |
| 337 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
-0.322 |
-0.117 |
| 338 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.322 |
-0.048 |
| 339 |
265704_at |
AT2G03420
|
unknown |
-0.322 |
-0.021 |
| 340 |
259207_at |
AT3G09050
|
unknown |
-0.321 |
-0.039 |
| 341 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.321 |
-0.019 |
| 342 |
262342_at |
AT1G64150
|
[Uncharacterized protein family (UPF0016)] |
-0.320 |
-0.011 |
| 343 |
248920_at |
AT5G45930
|
CHL I2, CHLI-2, CHLI2, magnesium chelatase i2 |
-0.320 |
0.009 |
| 344 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.320 |
0.024 |
| 345 |
255793_at |
AT2G33250
|
unknown |
-0.319 |
-0.055 |
| 346 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.319 |
0.084 |
| 347 |
248380_at |
AT5G51820
|
ATPGMP, ARABIDOPSIS THALIANA PHOSPHOGLUCOMUTASE, PGM, phosphoglucomutase, PGM1, STF1, STARCH-FREE 1 |
-0.317 |
0.010 |
| 348 |
255623_at |
AT4G01310
|
[Ribosomal L5P family protein] |
-0.316 |
-0.010 |
| 349 |
264371_at |
AT1G12090
|
ELP, extensin-like protein |
-0.316 |
-0.019 |
| 350 |
267299_at |
AT2G30150
|
[UDP-Glycosyltransferase superfamily protein] |
-0.316 |
-0.084 |
| 351 |
266329_at |
AT2G01590
|
CRR3, CHLORORESPIRATORY REDUCTION 3 |
-0.314 |
-0.059 |
| 352 |
263942_at |
AT2G35860
|
FLA16, FASCICLIN-like arabinogalactan protein 16 precursor |
-0.313 |
-0.035 |
| 353 |
249230_at |
AT5G42070
|
unknown |
-0.313 |
-0.052 |
| 354 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.312 |
-0.040 |
| 355 |
256746_at |
AT3G29320
|
PHS1, alpha-glucan phosphorylase 1 |
-0.312 |
0.065 |
| 356 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
-0.312 |
-0.002 |
| 357 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
-0.311 |
0.025 |
| 358 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.310 |
0.017 |
| 359 |
251008_at |
AT5G02710
|
unknown |
-0.310 |
-0.030 |
| 360 |
260106_at |
AT1G35420
|
[alpha/beta-Hydrolases superfamily protein] |
-0.310 |
-0.005 |
| 361 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
-0.309 |
-0.047 |
| 362 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.308 |
-0.001 |
| 363 |
255016_at |
AT4G10120
|
ATSPS4F |
-0.308 |
-0.198 |
| 364 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.308 |
-0.039 |
| 365 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
-0.307 |
-0.013 |
| 366 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.302 |
-0.053 |
| 367 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
-0.302 |
-0.120 |
| 368 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.300 |
-0.040 |
| 369 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.299 |
-0.057 |
| 370 |
248287_at |
AT5G52970
|
[thylakoid lumen 15.0 kDa protein] |
-0.299 |
-0.006 |
| 371 |
248242_at |
AT5G53580
|
AtPLR1, PLR1, pyridoxal reductase 1 |
-0.299 |
-0.055 |
| 372 |
262897_at |
AT1G59840
|
CCB4, cofactor assembly of complex C |
-0.299 |
-0.009 |
| 373 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.298 |
0.029 |
| 374 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.298 |
-0.008 |
| 375 |
260368_at |
AT1G69700
|
ATHVA22C, HVA22 homologue C, HVA22C, HVA22 homologue C |
-0.297 |
-0.002 |
| 376 |
250906_at |
AT5G03650
|
SBE2.2, starch branching enzyme 2.2 |
-0.297 |
-0.029 |
| 377 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.296 |
-0.015 |
| 378 |
256455_at |
AT1G75190
|
unknown |
-0.295 |
0.131 |
| 379 |
254982_at |
AT4G10470
|
unknown |
-0.295 |
-0.032 |
| 380 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.295 |
-0.011 |
| 381 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.294 |
-0.063 |
| 382 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.294 |
-0.018 |
| 383 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.293 |
-0.100 |
| 384 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.292 |
-0.041 |
| 385 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.291 |
-0.003 |
| 386 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.290 |
-0.159 |
| 387 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.289 |
0.047 |
| 388 |
261084_at |
AT1G07440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.288 |
-0.047 |
| 389 |
248409_at |
AT5G51545
|
LPA2, low psii accumulation2 |
-0.288 |
0.013 |
| 390 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.287 |
-0.032 |
| 391 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
-0.287 |
-0.110 |
| 392 |
251670_at |
AT3G57190
|
PrfB3, peptide chain release factor 3 |
-0.287 |
-0.042 |
| 393 |
263287_at |
AT2G36145
|
unknown |
-0.285 |
-0.021 |
| 394 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.285 |
-0.044 |
| 395 |
261507_at |
AT1G71720
|
PDE338, PIGMENT DEFECTIVE 338, RLSB, RbcL mRNA S1 binding domain protein |
-0.284 |
-0.011 |
| 396 |
253876_at |
AT4G27430
|
CIP7, COP1-interacting protein 7 |
-0.284 |
-0.057 |
| 397 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
-0.284 |
-0.011 |
| 398 |
257236_at |
AT3G15095
|
HCF243, high chlorophyll fluorescence 243 |
-0.283 |
-0.006 |
| 399 |
255436_at |
AT4G03150
|
unknown |
-0.282 |
-0.037 |
| 400 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.281 |
-0.031 |
| 401 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.281 |
-0.108 |
| 402 |
248962_at |
AT5G45680
|
ATFKBP13, FK506 BINDING PROTEIN 13, FKBP13, FK506-binding protein 13 |
-0.281 |
-0.025 |
| 403 |
249118_at |
AT5G43870
|
[FUNCTIONS IN: phosphoinositide binding] |
-0.281 |
-0.125 |
| 404 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.280 |
0.003 |
| 405 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
-0.280 |
-0.015 |
| 406 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.280 |
-0.083 |
| 407 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
-0.280 |
-0.038 |
| 408 |
248669_at |
AT5G48730
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.279 |
-0.116 |
| 409 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
-0.278 |
-0.033 |
| 410 |
259132_at |
AT3G02250
|
[O-fucosyltransferase family protein] |
-0.278 |
-0.038 |
| 411 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.278 |
-0.060 |
| 412 |
245492_at |
AT4G16340
|
SPK1, SPIKE1 |
-0.277 |
0.034 |
| 413 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.277 |
0.027 |
| 414 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.277 |
0.341 |
| 415 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
-0.277 |
-0.015 |
| 416 |
256396_at |
AT3G06150
|
unknown |
-0.276 |
-0.050 |
| 417 |
250985_at |
AT5G02830
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.276 |
-0.134 |
| 418 |
261577_at |
AT1G01080
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.276 |
0.002 |
| 419 |
251083_at |
AT5G01590
|
TIC56, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 56 |
-0.276 |
-0.009 |
| 420 |
251243_at |
AT3G61870
|
unknown |
-0.276 |
-0.026 |
| 421 |
260696_at |
AT1G32520
|
unknown |
-0.275 |
-0.118 |
| 422 |
247486_at |
AT5G62140
|
unknown |
-0.275 |
-0.100 |
| 423 |
260837_at |
AT1G43670
|
AtcFBP, Arabidopsis thaliana cytosolic fructose-1,6-bisphosphatase, FBP, fructose-1,6-bisphosphatase, FINS1, FRUCTOSE INSENSITIVE 1 |
-0.275 |
-0.035 |
| 424 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.275 |
-0.044 |
| 425 |
247826_at |
AT5G58480
|
[O-Glycosyl hydrolases family 17 protein] |
-0.274 |
0.000 |
| 426 |
245347_at |
AT4G14890
|
FdC1, ferredoxin C 1 |
-0.274 |
-0.027 |
| 427 |
263678_at |
AT1G04420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.273 |
-0.029 |
| 428 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.273 |
-0.120 |
| 429 |
245952_at |
AT5G28500
|
unknown |
-0.272 |
-0.019 |
| 430 |
258156_at |
AT3G18050
|
unknown |
-0.272 |
-0.031 |
| 431 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.272 |
-0.116 |
| 432 |
254508_at |
AT4G20170
|
GALS3, galactan synthase 3 |
-0.272 |
-0.070 |
| 433 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
-0.271 |
0.042 |
| 434 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
-0.271 |
-0.013 |
| 435 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
-0.271 |
-0.053 |
| 436 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.270 |
-0.002 |
| 437 |
260523_at |
AT2G41720
|
EMB2654, EMBRYO DEFECTIVE 2654 |
-0.270 |
-0.024 |
| 438 |
261078_at |
AT1G07320
|
EMB2784, EMBRYO DEFECTIVE 2784, PRPL4, plastid ribosomal protein L4, RPL4, ribosomal protein L4 |
-0.270 |
-0.009 |
| 439 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.269 |
-0.038 |
| 440 |
258641_at |
AT3G08030
|
unknown |
-0.269 |
0.033 |
| 441 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
-0.268 |
-0.007 |