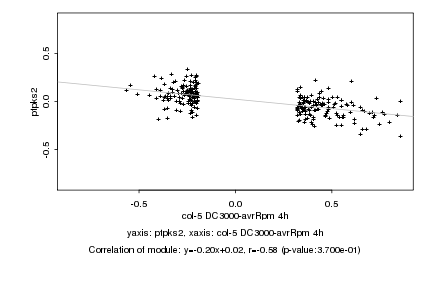
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
col-5 DC3000-avrRpm 4h |
Log2 signal ratio
ptpks2 |
| 1 |
248090_at |
AT5G55090
|
MAPKKK15, mitogen-activated protein kinase kinase kinase 15 |
0.854 |
0.003 |
| 2 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
0.851 |
-0.355 |
| 3 |
253044_at |
AT4G37290
|
unknown |
0.840 |
-0.137 |
| 4 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.796 |
-0.209 |
| 5 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.770 |
-0.130 |
| 6 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
0.760 |
-0.112 |
| 7 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.744 |
-0.230 |
| 8 |
247064_at |
AT5G66890
|
[Leucine-rich repeat (LRR) family protein] |
0.730 |
0.040 |
| 9 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.718 |
-0.136 |
| 10 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.714 |
-0.156 |
| 11 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.708 |
-0.109 |
| 12 |
266010_at |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
0.694 |
-0.124 |
| 13 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.679 |
-0.290 |
| 14 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
0.668 |
-0.099 |
| 15 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
0.658 |
-0.287 |
| 16 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.655 |
-0.088 |
| 17 |
263455_at |
AT2G22320
|
unknown |
0.645 |
-0.342 |
| 18 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.644 |
-0.062 |
| 19 |
251334_at |
AT3G61390
|
[RING/U-box superfamily protein] |
0.614 |
-0.185 |
| 20 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.614 |
-0.227 |
| 21 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.612 |
-0.039 |
| 22 |
249896_at |
AT5G22530
|
unknown |
0.601 |
0.209 |
| 23 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.597 |
-0.008 |
| 24 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.594 |
-0.108 |
| 25 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.581 |
-0.041 |
| 26 |
245738_at |
AT1G44130
|
[Eukaryotic aspartyl protease family protein] |
0.573 |
-0.025 |
| 27 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.560 |
-0.141 |
| 28 |
261429_at |
AT1G18860
|
ATWRKY61, WRKY DNA-BINDING PROTEIN 61, WRKY61, WRKY DNA-binding protein 61 |
0.559 |
-0.162 |
| 29 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
0.555 |
-0.157 |
| 30 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
0.550 |
0.015 |
| 31 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.550 |
-0.240 |
| 32 |
249263_at |
AT5G41730
|
[Protein kinase family protein] |
0.549 |
0.015 |
| 33 |
252345_at |
AT3G48640
|
unknown |
0.545 |
-0.050 |
| 34 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.538 |
-0.157 |
| 35 |
267451_at |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
0.528 |
-0.138 |
| 36 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.525 |
0.047 |
| 37 |
254347_at |
AT4G22070
|
ATWRKY31, WRKY31, WRKY DNA-binding protein 31 |
0.524 |
-0.246 |
| 38 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.520 |
-0.121 |
| 39 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.517 |
-0.012 |
| 40 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
0.509 |
-0.031 |
| 41 |
264646_at |
AT1G08860
|
BON3, BONZAI 3 |
0.508 |
-0.055 |
| 42 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
0.500 |
0.048 |
| 43 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
0.485 |
-0.176 |
| 44 |
246332_at |
AT3G44830
|
[Lecithin:cholesterol acyltransferase family protein] |
0.481 |
-0.058 |
| 45 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.481 |
0.140 |
| 46 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.480 |
0.022 |
| 47 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.479 |
-0.003 |
| 48 |
245447_at |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
0.479 |
-0.080 |
| 49 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.474 |
-0.097 |
| 50 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.469 |
-0.120 |
| 51 |
254288_at |
AT4G22970
|
AESP, homolog of separase, ESP, EXTRA SPINDLE POLES, ESP, homolog of separase, RSW4, RADIALLY SWOLLEN 4 |
0.465 |
-0.151 |
| 52 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.464 |
-0.136 |
| 53 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.458 |
-0.025 |
| 54 |
266779_at |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
0.454 |
-0.022 |
| 55 |
257583_at |
AT1G66480
|
[plastid movement impaired 2 (PMI2)] |
0.452 |
0.040 |
| 56 |
254897_at |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
0.450 |
-0.012 |
| 57 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.449 |
-0.041 |
| 58 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.444 |
-0.032 |
| 59 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.443 |
0.112 |
| 60 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.437 |
-0.030 |
| 61 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
0.433 |
0.043 |
| 62 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
0.431 |
0.089 |
| 63 |
260005_at |
AT1G67920
|
unknown |
0.429 |
-0.080 |
| 64 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.427 |
0.000 |
| 65 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.422 |
0.030 |
| 66 |
256370_at |
AT1G66780
|
[MATE efflux family protein] |
0.421 |
-0.078 |
| 67 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.417 |
-0.040 |
| 68 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.416 |
-0.029 |
| 69 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
0.415 |
0.219 |
| 70 |
261836_at |
AT1G16090
|
WAKL7, wall associated kinase-like 7 |
0.414 |
-0.089 |
| 71 |
250493_at |
AT5G09800
|
[ARM repeat superfamily protein] |
0.412 |
-0.007 |
| 72 |
248934_at |
AT5G46080
|
[Protein kinase superfamily protein] |
0.410 |
-0.250 |
| 73 |
247940_at |
AT5G57190
|
PSD2, phosphatidylserine decarboxylase 2 |
0.405 |
-0.092 |
| 74 |
252484_at |
AT3G46690
|
[UDP-Glycosyltransferase superfamily protein] |
0.404 |
-0.040 |
| 75 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
0.403 |
0.008 |
| 76 |
250415_at |
AT5G11210
|
ATGLR2.5, ARABIDOPSIS THALIANA GLU, GLR2.5, glutamate receptor 2.5 |
0.403 |
-0.186 |
| 77 |
264716_at |
AT1G70170
|
MMP, matrix metalloproteinase |
0.400 |
-0.165 |
| 78 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.398 |
-0.230 |
| 79 |
253819_at |
AT4G28350
|
LecRK-VII.2, L-type lectin receptor kinase VII.2 |
0.394 |
-0.212 |
| 80 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
0.394 |
-0.058 |
| 81 |
245082_at |
AT2G23270
|
unknown |
0.392 |
0.072 |
| 82 |
255884_at |
AT1G20310
|
unknown |
0.388 |
0.001 |
| 83 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
0.388 |
-0.144 |
| 84 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.383 |
-0.129 |
| 85 |
256499_at |
AT1G36640
|
unknown |
0.380 |
-0.128 |
| 86 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
0.380 |
-0.092 |
| 87 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.379 |
-0.016 |
| 88 |
267289_at |
AT2G23770
|
LYK4, LysM-containing receptor-like kinase 4 |
0.374 |
-0.080 |
| 89 |
266196_at |
AT2G39110
|
[Protein kinase superfamily protein] |
0.374 |
0.000 |
| 90 |
246406_at |
AT1G57650
|
[ATP binding] |
0.373 |
-0.174 |
| 91 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
0.372 |
0.049 |
| 92 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.365 |
0.016 |
| 93 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.365 |
0.010 |
| 94 |
260468_at |
AT1G11100
|
[SNF2 domain-containing protein / helicase domain-containing protein / zinc finger protein-related] |
0.361 |
-0.054 |
| 95 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.361 |
-0.098 |
| 96 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.359 |
-0.181 |
| 97 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.359 |
-0.132 |
| 98 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
0.357 |
-0.057 |
| 99 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.357 |
-0.090 |
| 100 |
255845_at |
AT2G33600
|
AtCRL2, CRL2, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 2 |
0.356 |
0.016 |
| 101 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.356 |
-0.215 |
| 102 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
0.354 |
0.052 |
| 103 |
250821_at |
AT5G05190
|
unknown |
0.354 |
0.050 |
| 104 |
254468_at |
AT4G20460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.352 |
-0.077 |
| 105 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.352 |
0.013 |
| 106 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.349 |
-0.001 |
| 107 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.348 |
-0.017 |
| 108 |
267032_at |
AT2G38490
|
CIPK22, CBL-interacting protein kinase 22, SnRK3.19, SNF1-RELATED PROTEIN KINASE 3.19 |
0.346 |
-0.053 |
| 109 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.344 |
0.042 |
| 110 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.344 |
-0.097 |
| 111 |
248848_at |
AT5G46520
|
VICTR, VARIATION IN COMPOUND TRIGGERED ROOT growth response |
0.339 |
-0.068 |
| 112 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
0.337 |
0.068 |
| 113 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.336 |
0.038 |
| 114 |
253012_at |
AT4G37900
|
unknown |
0.335 |
-0.045 |
| 115 |
261241_at |
AT1G32950
|
[Subtilase family protein] |
0.335 |
-0.129 |
| 116 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.334 |
0.154 |
| 117 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.333 |
-0.024 |
| 118 |
246790_at |
AT5G27610
|
ALY1, ALWAYS EARLY 1, ATALY1, ARABIDOPSIS THALIANA ALWAYS EARLY 1 |
0.332 |
-0.007 |
| 119 |
247618_at |
AT5G60280
|
LecRK-I.8, L-type lectin receptor kinase I.8 |
0.332 |
0.052 |
| 120 |
251689_at |
AT3G56500
|
[serine-rich protein-related] |
0.331 |
-0.113 |
| 121 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.329 |
-0.196 |
| 122 |
248936_at |
AT5G45710
|
AT-HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, HSFA4C, HEAT SHOCK TRANSCRIPTION FACTOR A4C, RHA1, ROOT HANDEDNESS 1 |
0.326 |
-0.034 |
| 123 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.325 |
-0.202 |
| 124 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.321 |
-0.141 |
| 125 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
0.320 |
-0.057 |
| 126 |
251906_at |
AT3G53720
|
ATCHX20, cation/H+ exchanger 20, CHX20, cation/H+ exchanger 20 |
0.320 |
-0.088 |
| 127 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.318 |
0.107 |
| 128 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.318 |
0.126 |
| 129 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.318 |
-0.063 |
| 130 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
0.318 |
-0.076 |
| 131 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.566 |
0.121 |
| 132 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.549 |
0.174 |
| 133 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.511 |
0.081 |
| 134 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.448 |
0.067 |
| 135 |
257076_at |
AT3G19680
|
unknown |
-0.420 |
0.261 |
| 136 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.413 |
0.126 |
| 137 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.412 |
0.036 |
| 138 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.400 |
-0.182 |
| 139 |
267423_at |
AT2G35060
|
KUP11, K+ uptake permease 11 |
-0.393 |
0.124 |
| 140 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.392 |
0.057 |
| 141 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.388 |
0.246 |
| 142 |
247601_at |
AT5G60850
|
OBP4, OBF binding protein 4 |
-0.378 |
0.017 |
| 143 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.372 |
0.178 |
| 144 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.372 |
0.049 |
| 145 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.371 |
-0.078 |
| 146 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.364 |
0.056 |
| 147 |
251673_at |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
-0.357 |
0.041 |
| 148 |
259038_at |
AT3G09210
|
PTAC13, plastid transcriptionally active 13 |
-0.356 |
0.054 |
| 149 |
247903_at |
AT5G57340
|
unknown |
-0.355 |
-0.169 |
| 150 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.354 |
-0.069 |
| 151 |
262230_at |
AT1G68560
|
ATXYL1, ALPHA-XYLOSIDASE 1, AXY3, altered xyloglucan 3, TRG1, thermoinhibition resistant germination 1, XYL1, alpha-xylosidase 1 |
-0.350 |
0.088 |
| 152 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.348 |
0.016 |
| 153 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.341 |
0.057 |
| 154 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.340 |
0.037 |
| 155 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.339 |
0.145 |
| 156 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.334 |
0.284 |
| 157 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.331 |
0.133 |
| 158 |
266481_at |
AT2G31070
|
TCP10, TCP domain protein 10 |
-0.328 |
0.095 |
| 159 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.326 |
0.207 |
| 160 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.317 |
0.061 |
| 161 |
258804_at |
AT3G04760
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.314 |
0.215 |
| 162 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.310 |
-0.089 |
| 163 |
259240_at |
AT3G11590
|
unknown |
-0.307 |
0.003 |
| 164 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.305 |
0.123 |
| 165 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.296 |
0.093 |
| 166 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.293 |
0.008 |
| 167 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.292 |
0.042 |
| 168 |
248773_at |
AT5G47820
|
FRA1, FRAGILE FIBER 1 |
-0.291 |
0.006 |
| 169 |
251753_at |
AT3G55760
|
unknown |
-0.286 |
-0.094 |
| 170 |
259017_at |
AT3G07310
|
unknown |
-0.286 |
-0.015 |
| 171 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
-0.282 |
0.090 |
| 172 |
263943_at |
AT2G35800
|
SAMTL, S-adenosyl methionine transporter-like |
-0.280 |
0.165 |
| 173 |
267532_at |
AT2G42040
|
unknown |
-0.277 |
0.039 |
| 174 |
253876_at |
AT4G27430
|
CIP7, COP1-interacting protein 7 |
-0.276 |
0.145 |
| 175 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.276 |
0.156 |
| 176 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.272 |
0.114 |
| 177 |
246990_at |
AT5G67360
|
ARA12 |
-0.271 |
0.176 |
| 178 |
251929_at |
AT3G53920
|
SIG3, SIGMA FACTOR 3, SIGC, RNApolymerase sigma-subunit C |
-0.270 |
-0.025 |
| 179 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
-0.269 |
0.073 |
| 180 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.267 |
0.221 |
| 181 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.261 |
0.079 |
| 182 |
251008_at |
AT5G02710
|
unknown |
-0.259 |
0.163 |
| 183 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-0.256 |
0.087 |
| 184 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.256 |
0.107 |
| 185 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.255 |
0.263 |
| 186 |
257722_at |
AT3G18490
|
ASPG1, ASPARTIC PROTEASE IN GUARD CELL 1 |
-0.254 |
0.087 |
| 187 |
247446_at |
AT5G62650
|
[Tic22-like family protein] |
-0.253 |
0.097 |
| 188 |
248780_at |
AT5G47760
|
ATPGLP2, 2-phosphoglycolate phosphatase 2, ATPK5, PGLP2, PGLP2, 2-phosphoglycolate phosphatase 2 |
-0.252 |
0.097 |
| 189 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.252 |
0.343 |
| 190 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
-0.249 |
0.115 |
| 191 |
258643_at |
AT3G08010
|
ATAB2 |
-0.248 |
-0.011 |
| 192 |
259140_at |
AT3G10230
|
AtLCY, LYC, lycopene cyclase |
-0.246 |
0.003 |
| 193 |
255948_at |
AT1G22060
|
[LOCATED IN: vacuole] |
-0.245 |
0.066 |
| 194 |
251719_at |
AT3G56140
|
unknown |
-0.244 |
-0.012 |
| 195 |
245354_at |
AT4G17600
|
LIL3:1 |
-0.243 |
0.030 |
| 196 |
250865_at |
AT5G03900
|
[Iron-sulphur cluster biosynthesis family protein] |
-0.243 |
0.071 |
| 197 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.243 |
0.113 |
| 198 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.240 |
0.062 |
| 199 |
250495_at |
AT5G09770
|
[Ribosomal protein L17 family protein] |
-0.238 |
-0.011 |
| 200 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.238 |
-0.115 |
| 201 |
251936_at |
AT3G53700
|
MEE40, maternal effect embryo arrest 40 |
-0.238 |
0.130 |
| 202 |
257937_at |
AT3G19810
|
unknown |
-0.236 |
0.141 |
| 203 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
-0.236 |
0.169 |
| 204 |
253989_at |
AT4G26130
|
unknown |
-0.236 |
0.216 |
| 205 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
-0.234 |
0.076 |
| 206 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.234 |
0.023 |
| 207 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.233 |
0.273 |
| 208 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
-0.232 |
0.011 |
| 209 |
256825_at |
AT3G22120
|
CWLP, cell wall-plasma membrane linker protein |
-0.231 |
-0.032 |
| 210 |
251500_at |
AT3G59110
|
[Protein kinase superfamily protein] |
-0.231 |
0.103 |
| 211 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.231 |
-0.086 |
| 212 |
258663_at |
AT3G08670
|
unknown |
-0.229 |
0.132 |
| 213 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
-0.228 |
0.107 |
| 214 |
267061_at |
AT2G32480
|
ARASP, ARABIDOPSIS SERIN PROTEASE |
-0.227 |
0.052 |
| 215 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
-0.225 |
-0.164 |
| 216 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.225 |
0.134 |
| 217 |
245396_at |
AT4G14870
|
SECE1 |
-0.225 |
-0.015 |
| 218 |
250441_at |
AT5G10540
|
TOP2, thimet metalloendopeptidase 2 |
-0.224 |
-0.016 |
| 219 |
257660_at |
AT3G13360
|
WIP3, WPP domain interacting protein 3 |
-0.224 |
-0.111 |
| 220 |
255793_at |
AT2G33250
|
unknown |
-0.222 |
0.089 |
| 221 |
258750_at |
AT3G05910
|
[Pectinacetylesterase family protein] |
-0.220 |
0.067 |
| 222 |
264781_at |
AT1G08540
|
ABC1, ATSIG1, SIGMA FACTOR 1, ATSIG2, SIGMA FACTOR 2, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, SIGB, SIGMA FACTOR B |
-0.218 |
0.205 |
| 223 |
248303_at |
AT5G53170
|
FTSH11, FTSH protease 11 |
-0.217 |
0.137 |
| 224 |
262727_at |
AT1G75800
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.217 |
-0.060 |
| 225 |
250430_at |
AT5G10460
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.216 |
0.059 |
| 226 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.214 |
-0.052 |
| 227 |
255047_at |
AT4G09670
|
[Oxidoreductase family protein] |
-0.213 |
0.040 |
| 228 |
258250_at |
AT3G15850
|
ADS3, AtADS3, FAD5, fatty acid desaturase 5, FADB, FATTY ACID DESATURASE B, JB67 |
-0.213 |
0.005 |
| 229 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.212 |
0.258 |
| 230 |
259505_at |
AT1G15810
|
[S15/NS1, RNA-binding protein] |
-0.211 |
0.171 |
| 231 |
263014_at |
AT1G23400
|
ATCAF2, ARABIDOPSIS THALIANA HOMOLOG OF MAIZE CAF2, CAF2 |
-0.211 |
-0.004 |
| 232 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
-0.211 |
0.107 |
| 233 |
254596_at |
AT4G18975
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.210 |
0.161 |
| 234 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.209 |
0.063 |
| 235 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
-0.209 |
0.000 |
| 236 |
248529_at |
AT5G50000
|
[Protein kinase superfamily protein] |
-0.208 |
0.224 |
| 237 |
252255_at |
AT3G49220
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.207 |
0.276 |
| 238 |
250444_at |
AT5G10560
|
[Glycosyl hydrolase family protein] |
-0.207 |
0.064 |
| 239 |
251959_at |
AT3G53410
|
LUL2, LOG2-LIKE UBIQUITIN LIGASE2 |
-0.207 |
0.145 |
| 240 |
262951_at |
AT1G75500
|
UMAMIT5, Usually multiple acids move in and out Transporters 5, WAT1, Walls Are Thin 1 |
-0.205 |
0.179 |
| 241 |
245165_at |
AT2G33180
|
unknown |
-0.204 |
0.089 |
| 242 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.204 |
0.202 |
| 243 |
261646_at |
AT1G27690
|
unknown |
-0.204 |
0.070 |
| 244 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.204 |
-0.140 |
| 245 |
250126_at |
AT5G16310
|
UCH1 |
-0.204 |
0.081 |
| 246 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.203 |
-0.016 |
| 247 |
258263_at |
AT3G15780
|
unknown |
-0.203 |
0.099 |
| 248 |
254356_at |
AT4G22190
|
unknown |
-0.202 |
0.082 |
| 249 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.202 |
0.270 |
| 250 |
264315_at |
AT1G70370
|
PG2, polygalacturonase 2 |
-0.201 |
0.019 |
| 251 |
260877_at |
AT1G21500
|
unknown |
-0.201 |
0.043 |
| 252 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
-0.201 |
-0.067 |
| 253 |
264799_at |
AT1G08550
|
AVDE1, ARABIDOPSIS VIOLAXANTHIN DE-EPOXIDASE 1, NPQ1, non-photochemical quenching 1 |
-0.199 |
0.049 |
| 254 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
-0.198 |
0.021 |
| 255 |
248721_at |
AT5G47780
|
GAUT4, galacturonosyltransferase 4 |
-0.198 |
0.099 |
| 256 |
258161_at |
AT3G17930
|
DAC, Defective Accumulation of Cytochrome b6/f complex |
-0.198 |
0.043 |
| 257 |
250133_at |
AT5G16400
|
ATF2, TRXF2, thioredoxin F2 |
-0.197 |
0.001 |
| 258 |
260271_at |
AT1G63690
|
ATSPPL2, SIGNAL PEPTIDE PEPTIDASE-LIKE 2, SPPL2, SIGNAL PEPTIDE PEPTIDASE-LIKE 2 |
-0.196 |
0.072 |
| 259 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.194 |
0.195 |