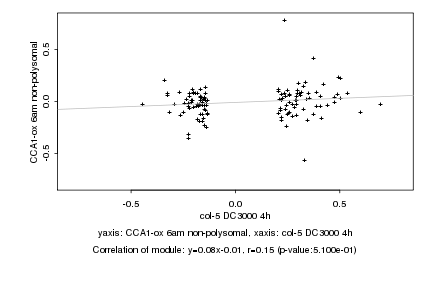
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
col-5 DC3000 4h |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
256181_at |
AT1G51820
|
[Leucine-rich repeat protein kinase family protein] |
0.692 |
-0.021 |
| 2 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.597 |
-0.103 |
| 3 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.533 |
0.083 |
| 4 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.503 |
0.230 |
| 5 |
256378_at |
AT1G66830
|
[Leucine-rich repeat protein kinase family protein] |
0.499 |
0.036 |
| 6 |
267588_at |
AT2G42060
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.492 |
0.233 |
| 7 |
266779_at |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
0.485 |
0.072 |
| 8 |
248700_at |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
0.472 |
0.043 |
| 9 |
263797_at |
AT2G24570
|
ATWRKY17, WRKY17, WRKY DNA-binding protein 17 |
0.470 |
-0.003 |
| 10 |
261006_at |
AT1G26410
|
[FAD-binding Berberine family protein] |
0.437 |
-0.037 |
| 11 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.420 |
0.171 |
| 12 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.408 |
-0.160 |
| 13 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.405 |
-0.044 |
| 14 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.405 |
0.053 |
| 15 |
245556_at |
AT4G15400
|
ABS1, ABNORMAL SHOOT 1, BIA1, BRASSINOSTEROID INACTIVATOR1 |
0.386 |
-0.041 |
| 16 |
260432_at |
AT1G68150
|
ATWRKY9, WRKY9, WRKY DNA-binding protein 9 |
0.385 |
0.089 |
| 17 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.372 |
0.422 |
| 18 |
248980_at |
AT5G45090
|
AtPP2-A7, phloem protein 2-A7, PP2-A7, phloem protein 2-A7 |
0.371 |
-0.124 |
| 19 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
0.351 |
0.033 |
| 20 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.347 |
0.078 |
| 21 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.345 |
-0.182 |
| 22 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.339 |
0.026 |
| 23 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.333 |
0.186 |
| 24 |
254468_at |
AT4G20460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.327 |
-0.564 |
| 25 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.326 |
-0.076 |
| 26 |
255884_at |
AT1G20310
|
unknown |
0.323 |
0.148 |
| 27 |
250821_at |
AT5G05190
|
unknown |
0.314 |
0.096 |
| 28 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.310 |
0.067 |
| 29 |
248701_at |
AT5G48410
|
ATGLR1.3, ARABIDOPSIS THALIANA GLUTAMATE RECEPTOR 1.3, GLR1.3, glutamate receptor 1.3 |
0.305 |
0.086 |
| 30 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
0.298 |
0.179 |
| 31 |
263937_at |
AT2G35910
|
[RING/U-box superfamily protein] |
0.295 |
0.107 |
| 32 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.293 |
0.082 |
| 33 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
0.292 |
-0.023 |
| 34 |
256169_at |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
0.290 |
0.010 |
| 35 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.290 |
-0.125 |
| 36 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.289 |
0.051 |
| 37 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.284 |
0.010 |
| 38 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.282 |
-0.051 |
| 39 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.272 |
-0.138 |
| 40 |
258323_at |
AT3G22750
|
[Protein kinase superfamily protein] |
0.270 |
-0.027 |
| 41 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.259 |
-0.101 |
| 42 |
264357_at |
AT1G03360
|
ATRRP4, ribosomal RNA processing 4, RRP4, ribosomal RNA processing 4 |
0.258 |
-0.005 |
| 43 |
256499_at |
AT1G36640
|
unknown |
0.258 |
0.068 |
| 44 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.255 |
0.058 |
| 45 |
256158_at |
AT1G13590
|
ATPSK1, phytosulfokine 1 precursor, PSK1, phytosulfokine 1 precursor |
0.251 |
-0.111 |
| 46 |
247532_at |
AT5G61560
|
[U-box domain-containing protein kinase family protein] |
0.249 |
0.113 |
| 47 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
0.248 |
-0.119 |
| 48 |
247297_at |
AT5G64100
|
[Peroxidase superfamily protein] |
0.243 |
-0.237 |
| 49 |
257619_at |
AT3G24810
|
ICK3, KRP5, kip-related protein 5 |
0.241 |
-0.034 |
| 50 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
0.239 |
-0.071 |
| 51 |
247227_at |
AT5G65130
|
[Integrase-type DNA-binding superfamily protein] |
0.238 |
0.052 |
| 52 |
253493_at |
AT4G31820
|
ENP, ENHANCER OF PINOID, MAB4, MACCHI-BOU 4, NPY1, NAKED PINS IN YUC MUTANTS 1 |
0.237 |
0.050 |
| 53 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
0.234 |
0.085 |
| 54 |
258752_at |
AT3G09520
|
ATEXO70H4, exocyst subunit exo70 family protein H4, EXO70H4, exocyst subunit exo70 family protein H4 |
0.233 |
0.789 |
| 55 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.222 |
0.031 |
| 56 |
249943_at |
AT5G22280
|
unknown |
0.220 |
-0.147 |
| 57 |
246146_at |
AT5G20050
|
[Protein kinase superfamily protein] |
0.220 |
0.019 |
| 58 |
257377_at |
AT2G28890
|
PLL4, poltergeist like 4 |
0.217 |
-0.180 |
| 59 |
267226_at |
AT2G44010
|
unknown |
0.217 |
0.076 |
| 60 |
258396_at |
AT3G15460
|
[Ribosomal RNA processing Brix domain protein] |
0.215 |
-0.085 |
| 61 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.212 |
-0.059 |
| 62 |
267547_at |
AT2G32670
|
ATVAMP725, vesicle-associated membrane protein 725, VAMP725, vesicle-associated membrane protein 725 |
0.210 |
0.023 |
| 63 |
253012_at |
AT4G37900
|
unknown |
0.206 |
0.106 |
| 64 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.206 |
0.120 |
| 65 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.205 |
-0.106 |
| 66 |
250257_at |
AT5G13770
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
-0.447 |
-0.021 |
| 67 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.342 |
0.209 |
| 68 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.330 |
0.058 |
| 69 |
258113_at |
AT3G14650
|
CYP72A11, cytochrome P450, family 72, subfamily A, polypeptide 11 |
-0.328 |
0.080 |
| 70 |
263473_at |
AT2G31750
|
UGT74D1, UDP-glucosyl transferase 74D1 |
-0.320 |
-0.098 |
| 71 |
257076_at |
AT3G19680
|
unknown |
-0.296 |
-0.024 |
| 72 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.271 |
0.090 |
| 73 |
264247_at |
AT1G60160
|
[Potassium transporter family protein] |
-0.266 |
-0.129 |
| 74 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.249 |
-0.100 |
| 75 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.245 |
-0.012 |
| 76 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
-0.237 |
0.020 |
| 77 |
247248_at |
AT5G64560
|
ATMGT9, MGT9, magnesium transporter 9, MRS2-2 |
-0.229 |
-0.308 |
| 78 |
254785_at |
AT4G12730
|
FLA2, FASCICLIN-like arabinogalactan 2 |
-0.229 |
-0.355 |
| 79 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.227 |
-0.039 |
| 80 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.225 |
-0.017 |
| 81 |
246510_at |
AT5G15410
|
ATCNGC2, CYCLIC NUCLEOTIDE-GATED CHANNEL 2, CNGC2, CYCLIC NUCLEOTIDE GATED CHANNEL 2, DND1, DEFENSE NO DEATH 1 |
-0.223 |
-0.065 |
| 82 |
251500_at |
AT3G59110
|
[Protein kinase superfamily protein] |
-0.223 |
0.082 |
| 83 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.222 |
0.055 |
| 84 |
253876_at |
AT4G27430
|
CIP7, COP1-interacting protein 7 |
-0.214 |
-0.005 |
| 85 |
266990_at |
AT2G39190
|
ATATH8 |
-0.212 |
0.010 |
| 86 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.208 |
0.119 |
| 87 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.206 |
0.018 |
| 88 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-0.204 |
0.086 |
| 89 |
264781_at |
AT1G08540
|
ABC1, ATSIG1, SIGMA FACTOR 1, ATSIG2, SIGMA FACTOR 2, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, SIGB, SIGMA FACTOR B |
-0.203 |
-0.049 |
| 90 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
-0.202 |
0.093 |
| 91 |
246800_at |
AT5G26780
|
SHM2, serine hydroxymethyltransferase 2 |
-0.195 |
0.083 |
| 92 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.191 |
-0.042 |
| 93 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.190 |
-0.041 |
| 94 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.186 |
-0.164 |
| 95 |
259017_at |
AT3G07310
|
unknown |
-0.184 |
0.079 |
| 96 |
258663_at |
AT3G08670
|
unknown |
-0.184 |
-0.021 |
| 97 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.181 |
-0.033 |
| 98 |
252996_s_at |
AT4G38460
|
AtSSU, GGR, geranylgeranyl reductase, SSU, small subunit of Heterodimeric geranyl(geranyl)diphosphate synthase |
-0.179 |
-0.047 |
| 99 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
-0.175 |
-0.186 |
| 100 |
248303_at |
AT5G53170
|
FTSH11, FTSH protease 11 |
-0.173 |
-0.020 |
| 101 |
247861_at |
AT5G58160
|
[actin binding] |
-0.171 |
0.047 |
| 102 |
258750_at |
AT3G05910
|
[Pectinacetylesterase family protein] |
-0.171 |
0.056 |
| 103 |
256140_at |
AT1G48650
|
[DEA(D/H)-box RNA helicase family protein] |
-0.170 |
0.116 |
| 104 |
250250_at |
AT5G13610
|
unknown |
-0.168 |
0.011 |
| 105 |
258696_at |
AT3G09650
|
CRM3, HCF152, HIGH CHLOROPHYLL FLUORESCENCE 152 |
-0.168 |
-0.117 |
| 106 |
246829_at |
AT5G26570
|
ATGWD3, OK1, PWD, PHOSPHOGLUCAN WATER DIKINASE |
-0.164 |
0.045 |
| 107 |
247092_at |
AT5G66380
|
ATFOLT1, folate transporter 1, FOLT1, folate transporter 1 |
-0.162 |
-0.124 |
| 108 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.162 |
-0.030 |
| 109 |
255673_at |
AT4G00340
|
RLK4, receptor-like protein kinase 4 |
-0.160 |
-0.030 |
| 110 |
248560_at |
AT5G49970
|
pyridoxin (pyrodoxamine) 5'-phosphate oxidase |
-0.159 |
-0.006 |
| 111 |
250441_at |
AT5G10540
|
TOP2, thimet metalloendopeptidase 2 |
-0.159 |
-0.037 |
| 112 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.159 |
-0.189 |
| 113 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.157 |
-0.159 |
| 114 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
-0.156 |
0.023 |
| 115 |
260474_at |
AT1G11090
|
[alpha/beta-Hydrolases superfamily protein] |
-0.153 |
0.036 |
| 116 |
246990_at |
AT5G67360
|
ARA12 |
-0.153 |
-0.069 |
| 117 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.151 |
-0.227 |
| 118 |
246200_at |
AT4G37240
|
unknown |
-0.151 |
0.045 |
| 119 |
256754_at |
AT3G25690
|
AtCHUP1, Arabidopsis thaliana CHLOROPLAST UNUSUAL POSITIONING 1, CHUP1, CHLOROPLAST UNUSUAL POSITIONING 1 |
-0.150 |
-0.035 |
| 120 |
258151_at |
AT3G18080
|
BGLU44, B-S glucosidase 44 |
-0.147 |
0.085 |
| 121 |
246756_at |
AT5G27930
|
[Protein phosphatase 2C family protein] |
-0.145 |
-0.077 |
| 122 |
247186_at |
AT5G65470
|
[O-fucosyltransferase family protein] |
-0.144 |
0.019 |
| 123 |
266438_at |
AT2G43180
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.144 |
0.135 |
| 124 |
251068_at |
AT5G01920
|
STN8, State transition 8 |
-0.139 |
-0.030 |
| 125 |
266417_at |
AT2G38550
|
[Transmembrane proteins 14C] |
-0.139 |
-0.015 |
| 126 |
245776_at |
AT1G30260
|
unknown |
-0.139 |
-0.244 |
| 127 |
261909_at |
AT1G80680
|
MOS3, MODIFIER OF SNC1,3, NUP96, PRE, PRECOCIOUS, SAR3, SUPPRESSOR OF AUXIN RESISTANCE 3 |
-0.138 |
-0.118 |
| 128 |
250854_at |
AT5G04710
|
[Zn-dependent exopeptidases superfamily protein] |
-0.138 |
0.017 |
| 129 |
258291_at |
AT3G23310
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.137 |
-0.107 |