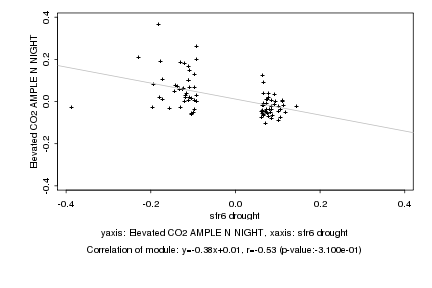
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sfr6 drought |
Log2 signal ratio
Elevated CO2 AMPLE N NIGHT |
| 1 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.143 |
-0.020 |
| 2 |
267637_at |
AT2G42190
|
unknown |
0.116 |
-0.051 |
| 3 |
251476_at |
AT3G59670
|
unknown |
0.113 |
-0.018 |
| 4 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.111 |
0.008 |
| 5 |
251418_at |
AT3G60440
|
[Phosphoglycerate mutase family protein] |
0.109 |
0.003 |
| 6 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
0.106 |
-0.075 |
| 7 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.105 |
-0.035 |
| 8 |
263192_at |
AT1G36160
|
ACC1, acetyl-CoA carboxylase 1, AT-ACC1, EMB22, EMBRYO DEFECTIVE 22, GK, GURKE, GSD1, GLOSSYHEAD 1, PAS3, PASTICCINO 3, SFR3, SENSITIVE TO FREEZING 3 |
0.101 |
-0.046 |
| 9 |
265656_at |
AT2G13820
|
AtXYP2, XYP2, xylogen protein 2 |
0.100 |
-0.022 |
| 10 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
0.100 |
-0.086 |
| 11 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
0.094 |
0.001 |
| 12 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.092 |
0.035 |
| 13 |
257730_at |
AT3G18420
|
[Protein prenylyltransferase superfamily protein] |
0.092 |
-0.013 |
| 14 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
0.086 |
-0.063 |
| 15 |
253817_at |
AT4G28310
|
unknown |
0.085 |
0.008 |
| 16 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.084 |
-0.076 |
| 17 |
252496_at |
AT3G46790
|
CRR2, CHLORORESPIRATORY REDUCTION 2 |
0.083 |
-0.019 |
| 18 |
249129_at |
AT5G43080
|
CYCA3;1, Cyclin A3;1 |
0.083 |
-0.036 |
| 19 |
253065_at |
AT4G37740
|
AtGRF2, growth-regulating factor 2, GRF2, growth-regulating factor 2 |
0.081 |
-0.052 |
| 20 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
0.080 |
-0.035 |
| 21 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.077 |
-0.068 |
| 22 |
263236_at |
AT1G10470
|
ARR4, response regulator 4, ATRR1, RESPONCE REGULATOR 1, IBC7, INDUCED BY CYTOKININ 7, MEE7, maternal effect embryo arrest 7 |
0.077 |
0.020 |
| 23 |
266959_at |
AT2G07690
|
MCM5, MINICHROMOSOME MAINTENANCE 5 |
0.076 |
0.040 |
| 24 |
266916_at |
AT2G45860
|
unknown |
0.075 |
-0.055 |
| 25 |
267307_at |
AT2G30210
|
LAC3, laccase 3 |
0.074 |
0.012 |
| 26 |
257660_at |
AT3G13360
|
WIP3, WPP domain interacting protein 3 |
0.073 |
-0.007 |
| 27 |
250434_at |
AT5G10390
|
H3.1, histone 3.1 |
0.072 |
0.014 |
| 28 |
265522_at |
AT2G06210
|
ELF8, EARLY FLOWERING 8, VIP6, VERNALIZATION INDEPENDENCE 6 |
0.072 |
-0.049 |
| 29 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.072 |
-0.035 |
| 30 |
250484_at |
AT5G10240
|
ASN3, asparagine synthetase 3 |
0.070 |
-0.047 |
| 31 |
258554_at |
AT3G06980
|
[DEA(D/H)-box RNA helicase family protein] |
0.069 |
-0.103 |
| 32 |
248066_at |
AT5G55590
|
QRT1, QUARTET 1 |
0.066 |
-0.061 |
| 33 |
260318_at |
AT1G63960
|
[Copper transport protein family] |
0.065 |
0.040 |
| 34 |
259735_at |
AT1G64405
|
unknown |
0.065 |
0.092 |
| 35 |
266980_at |
AT2G39390
|
[Ribosomal L29 family protein ] |
0.065 |
-0.017 |
| 36 |
261381_at |
AT1G05460
|
SDE3, SILENCING DEFECTIVE |
0.064 |
-0.063 |
| 37 |
247900_at |
AT5G57290
|
[60S acidic ribosomal protein family] |
0.064 |
-0.008 |
| 38 |
261130_at |
AT1G04870
|
ATPRMT10, PRMT10, protein arginine methyltransferase 10 |
0.063 |
-0.017 |
| 39 |
256666_at |
AT3G20670
|
HTA13, histone H2A 13 |
0.063 |
-0.043 |
| 40 |
257295_at |
AT3G17420
|
GPK1, glyoxysomal protein kinase 1 |
0.062 |
0.127 |
| 41 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
0.062 |
-0.054 |
| 42 |
257609_at |
AT3G13845
|
unknown |
0.062 |
-0.041 |
| 43 |
247556_at |
AT5G61040
|
unknown |
0.061 |
-0.045 |
| 44 |
257354_x_at |
AT2G23480
|
unknown |
0.060 |
-0.075 |
| 45 |
248937_at |
AT5G45770
|
AtRLP55, receptor like protein 55, RLP55, receptor like protein 55 |
-0.389 |
-0.027 |
| 46 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.231 |
0.209 |
| 47 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.196 |
-0.027 |
| 48 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.194 |
0.084 |
| 49 |
264580_at |
AT1G05340
|
unknown |
-0.182 |
0.366 |
| 50 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-0.181 |
0.020 |
| 51 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.179 |
0.194 |
| 52 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
-0.173 |
0.106 |
| 53 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.173 |
0.013 |
| 54 |
265387_at |
AT2G20670
|
unknown |
-0.157 |
-0.033 |
| 55 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.145 |
0.050 |
| 56 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
-0.142 |
0.078 |
| 57 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.137 |
0.075 |
| 58 |
261567_at |
AT1G33055
|
unknown |
-0.134 |
0.058 |
| 59 |
254571_at |
AT4G19370
|
unknown |
-0.131 |
0.186 |
| 60 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.130 |
-0.025 |
| 61 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
-0.126 |
0.058 |
| 62 |
255733_at |
AT1G25400
|
unknown |
-0.125 |
0.062 |
| 63 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
-0.121 |
0.185 |
| 64 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.121 |
0.002 |
| 65 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.120 |
0.023 |
| 66 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.119 |
0.031 |
| 67 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.117 |
0.042 |
| 68 |
258436_at |
AT3G16720
|
ATL2, TOXICOS EN LEVADURA 2, TL2, TOXICOS EN LEVADURA 2 |
-0.113 |
0.102 |
| 69 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.112 |
0.167 |
| 70 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.112 |
0.009 |
| 71 |
255243_at |
AT4G05590
|
unknown |
-0.111 |
0.104 |
| 72 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
-0.110 |
0.022 |
| 73 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.110 |
0.070 |
| 74 |
263565_at |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
-0.109 |
0.150 |
| 75 |
249234_at |
AT5G42200
|
[RING/U-box superfamily protein] |
-0.106 |
-0.061 |
| 76 |
249140_at |
AT5G43190
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.105 |
0.018 |
| 77 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.104 |
-0.053 |
| 78 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
-0.101 |
-0.052 |
| 79 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.099 |
0.009 |
| 80 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.099 |
0.130 |
| 81 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.099 |
-0.037 |
| 82 |
267381_at |
AT2G26190
|
[calmodulin-binding family protein] |
-0.097 |
0.067 |
| 83 |
252053_at |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
-0.094 |
0.202 |
| 84 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.094 |
0.002 |
| 85 |
253747_at |
AT4G29050
|
LecRK-V.9, L-type lectin receptor kinase V.9 |
-0.094 |
0.003 |
| 86 |
259479_at |
AT1G19020
|
unknown |
-0.093 |
0.263 |
| 87 |
262801_at |
AT1G21010
|
unknown |
-0.092 |
0.031 |