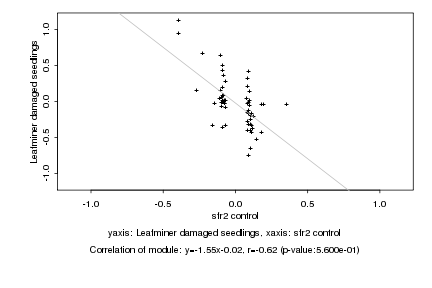
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
sfr2 control |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
254700_at |
AT4G18000
|
unknown |
0.353 |
-0.031 |
| 2 |
267560_at |
AT2G45580
|
CYP76C3, cytochrome P450, family 76, subfamily C, polypeptide 3 |
0.191 |
-0.032 |
| 3 |
259116_at |
AT3G01350
|
[Major facilitator superfamily protein] |
0.175 |
-0.424 |
| 4 |
254629_at |
AT4G18425
|
unknown |
0.174 |
-0.040 |
| 5 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
0.141 |
-0.524 |
| 6 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.119 |
-0.205 |
| 7 |
263545_at |
AT2G21560
|
unknown |
0.115 |
-0.365 |
| 8 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.108 |
-0.333 |
| 9 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
0.107 |
-0.417 |
| 10 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.105 |
-0.163 |
| 11 |
256673_at |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
0.101 |
-0.313 |
| 12 |
248753_at |
AT5G47630
|
mtACP3, mitochondrial acyl carrier protein 3 |
0.100 |
-0.189 |
| 13 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
0.100 |
-0.247 |
| 14 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
0.100 |
-0.650 |
| 15 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
0.098 |
-0.400 |
| 16 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.096 |
-0.054 |
| 17 |
253814_at |
AT4G28290
|
unknown |
0.092 |
0.149 |
| 18 |
245682_at |
AT5G08750
|
[RING/FYVE/PHD zinc finger superfamily protein] |
0.091 |
0.020 |
| 19 |
263782_at |
AT2G46380
|
unknown |
0.089 |
-0.312 |
| 20 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.085 |
-0.120 |
| 21 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
0.085 |
-0.748 |
| 22 |
250036_at |
AT5G18340
|
[ARM repeat superfamily protein] |
0.085 |
-0.163 |
| 23 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.084 |
0.421 |
| 24 |
260002_at |
AT1G67940
|
ABCI17, ATP-binding cassette I17, ATNAP3, non-intrinsic ABC protein 3, AtSTAR1, NAP3, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 3, NAP3, non-intrinsic ABC protein 3 |
0.084 |
-0.007 |
| 25 |
245800_at |
AT1G46264
|
AT-HSFB4, HSFB4, heat shock transcription factor B4, SCZ, SCHIZORIZA |
0.083 |
-0.146 |
| 26 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.083 |
-0.272 |
| 27 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.083 |
-0.391 |
| 28 |
248271_at |
AT5G53420
|
[CCT motif family protein] |
0.080 |
0.213 |
| 29 |
251062_at |
AT5G01840
|
ATOFP1, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1, OFP1, ovate family protein 1 |
0.078 |
0.324 |
| 30 |
251490_at |
AT3G59490
|
unknown |
0.077 |
-0.024 |
| 31 |
265460_at |
AT2G46600
|
[Calcium-binding EF-hand family protein] |
0.076 |
0.044 |
| 32 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
-0.398 |
1.140 |
| 33 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-0.398 |
0.951 |
| 34 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.274 |
0.155 |
| 35 |
252511_at |
AT3G46280
|
[protein kinase-related] |
-0.233 |
0.678 |
| 36 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.159 |
-0.321 |
| 37 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
-0.146 |
-0.026 |
| 38 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
-0.117 |
0.043 |
| 39 |
256324_at |
AT1G66760
|
[MATE efflux family protein] |
-0.109 |
0.640 |
| 40 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.107 |
0.164 |
| 41 |
264026_at |
AT2G21060
|
ATCSP4, COLD SHOCK DOMAIN PROTEIN 4, ATGRP2B, glycine-rich protein 2B, GRP2B, glycine-rich protein 2B |
-0.103 |
0.062 |
| 42 |
261117_at |
AT1G75310
|
AUL1, auxilin-like 1 |
-0.102 |
-0.061 |
| 43 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.101 |
-0.009 |
| 44 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.095 |
-0.348 |
| 45 |
264680_at |
AT1G65510
|
unknown |
-0.093 |
0.432 |
| 46 |
256910_at |
AT3G24080
|
[KRR1 family protein] |
-0.093 |
-0.007 |
| 47 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
-0.092 |
0.513 |
| 48 |
254364_at |
AT4G22020
|
[pseudogene] |
-0.092 |
0.021 |
| 49 |
259517_at |
AT1G20630
|
CAT1, catalase 1 |
-0.092 |
0.201 |
| 50 |
247888_at |
AT5G57920
|
AtENODL10, ENODL10, early nodulin-like protein 10 |
-0.091 |
0.087 |
| 51 |
260261_at |
AT1G68450
|
PDE337, PIGMENT DEFECTIVE 337 |
-0.089 |
0.009 |
| 52 |
256053_at |
AT1G07260
|
UGT71C3, UDP-glucosyl transferase 71C3 |
-0.088 |
0.364 |
| 53 |
262910_at |
AT1G59710
|
unknown |
-0.088 |
0.084 |
| 54 |
257342_at |
AT1G42365
|
[gypsy-like retrotransposon family (Athila), has a 9.0e-234 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.084 |
-0.013 |
| 55 |
260563_at |
AT2G43840
|
UGT74F1, UDP-glycosyltransferase 74 F1 |
-0.082 |
-0.018 |
| 56 |
251843_x_at |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
-0.081 |
-0.026 |
| 57 |
258014_at |
AT3G19330
|
unknown |
-0.076 |
0.019 |
| 58 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
-0.075 |
0.016 |
| 59 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.075 |
-0.328 |
| 60 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
-0.074 |
0.280 |
| 61 |
258704_at |
AT3G09780
|
ATCRR1, CCR1, CRINKLY4 related 1 |
-0.074 |
-0.071 |