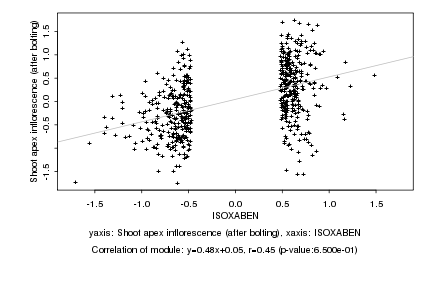
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ISOXABEN |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
245076_at |
AT2G23170
|
GH3.3 |
1.474 |
0.572 |
| 2 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
1.224 |
0.329 |
| 3 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
1.171 |
0.843 |
| 4 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
1.163 |
-0.383 |
| 5 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
1.151 |
-0.270 |
| 6 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
1.085 |
0.535 |
| 7 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.962 |
0.294 |
| 8 |
265076_at |
AT1G55490
|
CPN60B, chaperonin 60 beta, Cpn60beta1, chaperonin-60beta1, LEN1, LESION INITIATION 1 |
0.935 |
1.072 |
| 9 |
260242_at |
AT1G63650
|
AtEGL3, ATMYC-2, EGL1, EGL3, ENHANCER OF GLABRA 3 |
0.921 |
0.360 |
| 10 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
0.906 |
0.294 |
| 11 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
0.897 |
1.022 |
| 12 |
259727_at |
AT1G60950
|
ATFD2, FERREDOXIN 2, FD2, FED A |
0.893 |
1.016 |
| 13 |
265349_at |
AT2G22610
|
[Di-glucose binding protein with Kinesin motor domain] |
0.890 |
-0.097 |
| 14 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
0.868 |
1.637 |
| 15 |
252078_at |
AT3G51740
|
IMK2, inflorescence meristem receptor-like kinase 2 |
0.862 |
-0.081 |
| 16 |
259897_at |
AT1G71380
|
ATCEL3, cellulase 3, ATGH9B3, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3, CEL3, cellulase 3 |
0.858 |
-1.055 |
| 17 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
0.856 |
1.042 |
| 18 |
245688_at |
AT1G28290
|
AGP31, arabinogalactan protein 31 |
0.848 |
1.044 |
| 19 |
258369_at |
AT3G14310
|
ATPME3, pectin methylesterase 3, OZS2, OVERLY ZINC SENSITIVE 2, PME3, pectin methylesterase 3 |
0.848 |
0.720 |
| 20 |
255460_at |
AT4G02800
|
unknown |
0.841 |
1.255 |
| 21 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.840 |
0.133 |
| 22 |
246996_at |
AT5G67420
|
ASL39, ASYMMETRIC LEAVES2-LIKE 39, LBD37, LOB domain-containing protein 37 |
0.839 |
0.394 |
| 23 |
263607_at |
AT2G16270
|
unknown |
0.833 |
0.422 |
| 24 |
252148_at |
AT3G51280
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.825 |
1.107 |
| 25 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
0.824 |
0.292 |
| 26 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.817 |
1.536 |
| 27 |
262683_at |
AT1G75920
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.812 |
-1.155 |
| 28 |
261660_at |
AT1G18370
|
ATNACK1, ARABIDOPSIS NPK1-ACTIVATING KINESIN 1, HIK, HINKEL, NACK1, NPK1-ACTIVATING KINESIN 1 |
0.809 |
1.187 |
| 29 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.802 |
-0.935 |
| 30 |
248057_at |
AT5G55520
|
unknown |
0.797 |
0.386 |
| 31 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
0.796 |
0.777 |
| 32 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
0.796 |
0.797 |
| 33 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
0.795 |
1.098 |
| 34 |
248963_at |
AT5G45700
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.784 |
0.400 |
| 35 |
264312_at |
AT1G70450
|
[Protein kinase superfamily protein] |
0.781 |
-0.679 |
| 36 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
0.779 |
1.653 |
| 37 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.776 |
-0.652 |
| 38 |
266524_at |
AT2G16960
|
[ARM repeat superfamily protein] |
0.775 |
-0.864 |
| 39 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.769 |
-0.283 |
| 40 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
0.768 |
-0.205 |
| 41 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
0.765 |
0.593 |
| 42 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.764 |
1.031 |
| 43 |
263441_at |
AT2G28620
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.760 |
-0.375 |
| 44 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.753 |
1.169 |
| 45 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.750 |
-0.806 |
| 46 |
261368_at |
AT1G53070
|
[Legume lectin family protein] |
0.750 |
0.450 |
| 47 |
266305_at |
AT2G27120
|
POL2B, TIL2, TILTED 2 |
0.746 |
-0.693 |
| 48 |
266716_at |
AT2G46820
|
CURT1B, CURVATURE THYLAKOID 1B, PSAP, PSI-P, photosystem I P subunit, PTAC8, PLASTID TRANSCRIPTIONALLY ACTIVE 8, TMP14, THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA |
0.745 |
1.290 |
| 49 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
0.739 |
-0.698 |
| 50 |
245343_at |
AT4G15830
|
[ARM repeat superfamily protein] |
0.738 |
0.440 |
| 51 |
260131_at |
AT1G66310
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.733 |
-0.801 |
| 52 |
264545_at |
AT1G55670
|
PSAG, photosystem I subunit G |
0.729 |
0.828 |
| 53 |
264802_at |
AT1G08560
|
ATSYP111, KN, KNOLLE, SYP111, syntaxin of plants 111 |
0.722 |
0.270 |
| 54 |
249352_at |
AT5G40430
|
AtMYB22, myb domain protein 22, MYB22, myb domain protein 22 |
0.719 |
-1.548 |
| 55 |
245947_at |
AT5G19530
|
ACL5, ACAULIS 5 |
0.719 |
-0.181 |
| 56 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.714 |
0.332 |
| 57 |
267088_at |
AT2G38140
|
PSRP4, plastid-specific ribosomal protein 4 |
0.712 |
0.265 |
| 58 |
247425_at |
AT5G62550
|
unknown |
0.706 |
-0.162 |
| 59 |
261572_at |
AT1G01170
|
unknown |
0.703 |
1.435 |
| 60 |
246415_at |
AT5G17160
|
unknown |
0.702 |
0.654 |
| 61 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.700 |
-0.690 |
| 62 |
251846_at |
AT3G54560
|
HTA11, histone H2A 11 |
0.698 |
1.301 |
| 63 |
245347_at |
AT4G14890
|
FdC1, ferredoxin C 1 |
0.697 |
0.813 |
| 64 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
0.696 |
0.544 |
| 65 |
256899_at |
AT3G24660
|
TMKL1, transmembrane kinase-like 1 |
0.695 |
0.604 |
| 66 |
255623_at |
AT4G01310
|
[Ribosomal L5P family protein] |
0.695 |
1.260 |
| 67 |
257891_at |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
0.694 |
0.469 |
| 68 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
0.689 |
1.213 |
| 69 |
246557_at |
AT5G15510
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.687 |
-0.094 |
| 70 |
266172_at |
AT2G39010
|
PIP2;6, PLASMA MEMBRANE INTRINSIC PROTEIN 2;6, PIP2E, plasma membrane intrinsic protein 2E |
0.685 |
0.439 |
| 71 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.683 |
1.074 |
| 72 |
246344_at |
AT3G56730
|
[Putative endonuclease or glycosyl hydrolase] |
0.683 |
-0.829 |
| 73 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.681 |
0.843 |
| 74 |
246814_at |
AT5G27200
|
ACP5, acyl carrier protein 5 |
0.680 |
-1.271 |
| 75 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
0.678 |
1.687 |
| 76 |
266738_at |
AT2G47010
|
unknown |
0.673 |
0.651 |
| 77 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
0.671 |
1.335 |
| 78 |
267120_at |
AT2G23530
|
[Zinc-finger domain of monoamine-oxidase A repressor R1] |
0.669 |
0.034 |
| 79 |
245739_at |
AT1G44110
|
CYCA1;1, Cyclin A1;1 |
0.668 |
0.994 |
| 80 |
259930_at |
AT1G34355
|
ATPS1, PARALLEL SPINDLE 1, PS1, PARALLEL SPINDLE 1 |
0.667 |
-0.147 |
| 81 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.667 |
-1.088 |
| 82 |
256780_at |
AT3G13640
|
ABCE1, ATP-binding cassette E1, ATRLI1, RNAse l inhibitor protein 1, RLI1, RNAse l inhibitor protein 1 |
0.666 |
-0.230 |
| 83 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.666 |
-0.524 |
| 84 |
262081_at |
AT1G59540
|
ZCF125 |
0.665 |
0.005 |
| 85 |
251476_at |
AT3G59670
|
unknown |
0.663 |
-0.046 |
| 86 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
0.662 |
0.446 |
| 87 |
266655_at |
AT2G25880
|
AtAUR2, ataurora2, AUR2, ataurora2 |
0.661 |
0.661 |
| 88 |
257026_at |
AT3G19200
|
unknown |
0.659 |
0.183 |
| 89 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.657 |
1.251 |
| 90 |
247893_at |
AT5G57980
|
RPB5C, RNA polymerase II fifth largest subunit, C |
0.657 |
-0.325 |
| 91 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.656 |
0.300 |
| 92 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
0.654 |
-0.590 |
| 93 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
0.653 |
0.927 |
| 94 |
263640_at |
AT2G25270
|
unknown |
0.653 |
1.075 |
| 95 |
260934_at |
AT1G45100
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.652 |
-1.556 |
| 96 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
0.652 |
-0.523 |
| 97 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.651 |
0.384 |
| 98 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
0.651 |
0.142 |
| 99 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.650 |
0.099 |
| 100 |
248710_at |
AT5G48480
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.645 |
0.744 |
| 101 |
247450_at |
AT5G62350
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.642 |
0.626 |
| 102 |
258867_at |
AT3G03130
|
unknown |
0.641 |
0.139 |
| 103 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.640 |
0.327 |
| 104 |
252210_at |
AT3G50410
|
OBP1, OBF binding protein 1 |
0.638 |
0.682 |
| 105 |
264505_at |
AT1G09380
|
UMAMIT25, Usually multiple acids move in and out Transporters 25 |
0.638 |
-0.252 |
| 106 |
251483_at |
AT3G59650
|
[mitochondrial ribosomal protein L51/S25/CI-B8 family protein] |
0.637 |
0.971 |
| 107 |
244997_at |
ATCG00170
|
RPOC2 |
0.636 |
-0.076 |
| 108 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.635 |
1.102 |
| 109 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.634 |
0.226 |
| 110 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.633 |
0.508 |
| 111 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.633 |
0.517 |
| 112 |
265171_at |
AT1G23790
|
unknown |
0.633 |
-0.423 |
| 113 |
260481_at |
AT1G10960
|
ATFD1, ferredoxin 1, FD1, ferredoxin 1 |
0.632 |
0.381 |
| 114 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.631 |
1.297 |
| 115 |
265083_at |
AT1G03820
|
unknown |
0.629 |
0.175 |
| 116 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
0.628 |
0.253 |
| 117 |
248801_at |
AT5G47370
|
HAT2 |
0.627 |
0.153 |
| 118 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
0.627 |
-0.502 |
| 119 |
267636_at |
AT2G42110
|
unknown |
0.625 |
0.050 |
| 120 |
257115_at |
AT3G20150
|
[Kinesin motor family protein] |
0.623 |
-0.175 |
| 121 |
251566_at |
AT3G58210
|
[TRAF-like family protein] |
0.621 |
-0.692 |
| 122 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.621 |
0.589 |
| 123 |
254217_at |
AT4G23720
|
unknown |
0.620 |
0.372 |
| 124 |
258098_at |
AT3G23670
|
KINESIN-12B, PAKRP1L |
0.620 |
0.127 |
| 125 |
257807_at |
AT3G26650
|
GAPA, glyceraldehyde 3-phosphate dehydrogenase A subunit, GAPA-1, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT 1 |
0.620 |
1.757 |
| 126 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.619 |
0.617 |
| 127 |
261511_at |
AT1G71770
|
PAB5, poly(A)-binding protein 5 |
0.614 |
0.028 |
| 128 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
0.613 |
0.340 |
| 129 |
257773_at |
AT3G29185
|
unknown |
0.613 |
0.772 |
| 130 |
261460_at |
AT1G07880
|
ATMPK13 |
0.613 |
0.449 |
| 131 |
251982_at |
AT3G53190
|
[Pectin lyase-like superfamily protein] |
0.613 |
0.512 |
| 132 |
249381_at |
AT5G40040
|
[60S acidic ribosomal protein family] |
0.612 |
0.224 |
| 133 |
258067_at |
AT3G25980
|
AtMAD2, MAD2, MITOTIC ARREST-DEFICIENT 2 |
0.611 |
-0.080 |
| 134 |
264830_at |
AT1G03710
|
[Cystatin/monellin superfamily protein] |
0.611 |
0.251 |
| 135 |
245215_at |
AT1G67830
|
ATFXG1, Arabidopsis thaliana alpha-fucosidase 1, FXG1, alpha-fucosidase 1 |
0.611 |
0.772 |
| 136 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
0.605 |
0.054 |
| 137 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.605 |
0.017 |
| 138 |
264092_at |
AT1G79040
|
PSBR, photosystem II subunit R |
0.604 |
0.850 |
| 139 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
0.603 |
-0.198 |
| 140 |
263479_x_at |
AT2G04000
|
unknown |
0.600 |
-0.051 |
| 141 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.599 |
-0.148 |
| 142 |
254280_at |
AT4G22756
|
ATSMO1-2, SMO1-2, sterol C4-methyl oxidase 1-2 |
0.596 |
0.173 |
| 143 |
260553_at |
AT2G41800
|
unknown |
0.593 |
-0.304 |
| 144 |
252495_at |
AT3G46770
|
[AP2/B3-like transcriptional factor family protein] |
0.593 |
-1.010 |
| 145 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
0.593 |
0.604 |
| 146 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.590 |
0.678 |
| 147 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
0.590 |
1.362 |
| 148 |
258397_at |
AT3G15357
|
unknown |
0.590 |
-0.532 |
| 149 |
251735_at |
AT3G56090
|
ATFER3, ferritin 3, FER3, ferritin 3 |
0.588 |
0.433 |
| 150 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
0.587 |
0.631 |
| 151 |
253978_at |
AT4G26660
|
unknown |
0.586 |
0.421 |
| 152 |
251113_at |
AT5G01370
|
ACI1, ALC-interacting protein 1, TRM29, TON1 Recruiting Motif 29 |
0.586 |
0.147 |
| 153 |
252032_at |
AT3G52150
|
PSRP2, plastid-specific ribosomal protein 2 |
0.585 |
0.232 |
| 154 |
253771_at |
AT4G28430
|
[Reticulon family protein] |
0.583 |
-0.601 |
| 155 |
254623_at |
AT4G18480
|
CH-42, CHLORINA 42, CH42, CHLORINA 42, CHL11, CHLI-1, CHLI1 |
0.582 |
1.091 |
| 156 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.581 |
0.169 |
| 157 |
245044_at |
AT2G26500
|
[cytochrome b6f complex subunit (petM), putative] |
0.580 |
0.584 |
| 158 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
0.580 |
0.553 |
| 159 |
245165_at |
AT2G33180
|
unknown |
0.578 |
0.435 |
| 160 |
257140_at |
AT3G28910
|
ATMYB30, MYB30, myb domain protein 30 |
0.578 |
0.727 |
| 161 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
0.577 |
0.716 |
| 162 |
247320_at |
AT5G64040
|
PSAN |
0.576 |
1.067 |
| 163 |
264377_at |
AT2G25060
|
AtENODL14, ENODL14, early nodulin-like protein 14 |
0.575 |
0.674 |
| 164 |
265319_at |
AT2G22670
|
IAA8, indoleacetic acid-induced protein 8 |
0.574 |
0.794 |
| 165 |
263709_at |
AT1G09310
|
unknown |
0.574 |
0.145 |
| 166 |
263535_at |
AT2G24970
|
unknown |
0.574 |
0.723 |
| 167 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
0.574 |
0.966 |
| 168 |
257722_at |
AT3G18490
|
ASPG1, ASPARTIC PROTEASE IN GUARD CELL 1 |
0.571 |
0.914 |
| 169 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.571 |
-0.431 |
| 170 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.569 |
0.414 |
| 171 |
261055_at |
AT1G01300
|
[Eukaryotic aspartyl protease family protein] |
0.569 |
0.831 |
| 172 |
253090_at |
AT4G36360
|
BGAL3, beta-galactosidase 3 |
0.568 |
0.228 |
| 173 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.568 |
-0.905 |
| 174 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
0.564 |
1.143 |
| 175 |
255435_at |
AT4G03280
|
PETC, photosynthetic electron transfer C, PGR1, PROTON GRADIENT REGULATION 1 |
0.562 |
1.455 |
| 176 |
257929_at |
AT3G16980
|
NRPB9A, NRPD9A, NRPE9A |
0.562 |
0.837 |
| 177 |
249977_at |
AT5G18820
|
Cpn60alpha2, chaperonin-60alpha2, EMB3007, embryo defective 3007 |
0.562 |
-0.528 |
| 178 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
0.561 |
0.124 |
| 179 |
258087_at |
AT3G26060
|
ATPRX Q, PRXQ, peroxiredoxin Q |
0.561 |
0.577 |
| 180 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
0.560 |
1.038 |
| 181 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
0.560 |
0.891 |
| 182 |
260028_at |
AT1G29980
|
unknown |
0.557 |
0.371 |
| 183 |
245813_at |
AT1G49920
|
[MuDR family transposase] |
0.557 |
-0.941 |
| 184 |
264575_at |
AT1G05190
|
emb2394, embryo defective 2394 |
0.556 |
1.010 |
| 185 |
247376_at |
AT5G63310
|
ATNDPK2, ARABIDOPSIS NUCLEOSIDE DIPHOSPHATE KINASE 2, NDPK IA, NUCLEOSIDE DIPHOSPHATE KINASE IA, NDPK IA IA, NDPK1A, NDP KINASE 1A, NDPK2, nucleoside diphosphate kinase 2 |
0.555 |
1.368 |
| 186 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.554 |
0.832 |
| 187 |
256022_at |
AT1G58360
|
AAP1, amino acid permease 1, AtAAP1, NAT2, NEUTRAL AMINO ACID TRANSPORTER 2 |
0.554 |
0.114 |
| 188 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
0.554 |
0.786 |
| 189 |
256864_at |
AT3G23890
|
ATTOPII, TOPII, topoisomerase II |
0.554 |
0.357 |
| 190 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
0.553 |
0.006 |
| 191 |
252877_at |
AT4G39630
|
unknown |
0.553 |
0.783 |
| 192 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
0.551 |
1.020 |
| 193 |
256215_at |
AT1G50900
|
GDC1, Grana Deficient Chloroplast 1, LTD, LHCP translocation defect |
0.551 |
0.715 |
| 194 |
248186_at |
AT5G53880
|
unknown |
0.551 |
0.558 |
| 195 |
255410_at |
AT4G03100
|
[Rho GTPase activating protein with PAK-box/P21-Rho-binding domain] |
0.549 |
-0.076 |
| 196 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
0.549 |
0.328 |
| 197 |
262109_at |
AT1G02730
|
ATCSLD5, cellulose synthase-like D5, CSLD5, CELLULOSE SYNTHASE LIKE D5, CSLD5, cellulose synthase-like D5, SOS6, SALT OVERLY SENSITIVE 6 |
0.549 |
0.928 |
| 198 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
0.548 |
1.025 |
| 199 |
249832_at |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
0.547 |
-0.175 |
| 200 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.547 |
1.412 |
| 201 |
257831_at |
AT3G26710
|
CCB1, cofactor assembly of complex C |
0.546 |
0.763 |
| 202 |
256675_at |
AT3G52170
|
[DNA binding] |
0.546 |
-0.132 |
| 203 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
0.544 |
-1.457 |
| 204 |
265085_at |
AT1G03780
|
AtTPX2, TPX2, targeting protein for XKLP2 |
0.543 |
0.299 |
| 205 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
0.541 |
0.014 |
| 206 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.539 |
1.245 |
| 207 |
247948_at |
AT5G57130
|
SMXL5, SMAX1-like 5 |
0.539 |
-0.224 |
| 208 |
266598_at |
AT2G46110
|
KPHMT1, ketopantoate hydroxymethyltransferase 1, PANB1 |
0.539 |
-0.772 |
| 209 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.539 |
0.163 |
| 210 |
267612_at |
AT2G26690
|
AtNPF6.2, NPF6.2, NRT1/ PTR family 6.2 |
0.539 |
0.785 |
| 211 |
263736_at |
AT1G60000
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.538 |
0.483 |
| 212 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.537 |
0.719 |
| 213 |
267146_at |
AT2G38160
|
unknown |
0.536 |
0.604 |
| 214 |
260910_at |
AT1G02690
|
IMPA-6, importin alpha isoform 6 |
0.536 |
0.134 |
| 215 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.536 |
-0.319 |
| 216 |
259815_at |
AT1G49870
|
unknown |
0.535 |
0.146 |
| 217 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.535 |
-0.420 |
| 218 |
252442_at |
AT3G46940
|
DUT1, DUTP-PYROPHOSPHATASE-LIKE 1 |
0.535 |
0.457 |
| 219 |
260548_at |
AT2G43360
|
BIO2, BIOTIN AUXOTROPH 2, BIOB, BIOTIN AUXOTROPH B |
0.535 |
0.861 |
| 220 |
257815_at |
AT3G25130
|
unknown |
0.535 |
0.321 |
| 221 |
264352_at |
AT1G03270
|
unknown |
0.534 |
0.398 |
| 222 |
257054_at |
AT3G15353
|
ATMT3, MT3, metallothionein 3 |
0.532 |
0.583 |
| 223 |
252215_at |
AT3G50240
|
KICP-02 |
0.532 |
-0.361 |
| 224 |
247813_at |
AT5G58330
|
[lactate/malate dehydrogenase family protein] |
0.531 |
0.780 |
| 225 |
265870_at |
AT2G01660
|
PDLP6, plasmodesmata-located protein 6 |
0.531 |
-0.265 |
| 226 |
245357_at |
AT4G17560
|
[Ribosomal protein L19 family protein] |
0.530 |
-0.053 |
| 227 |
258641_at |
AT3G08030
|
unknown |
0.530 |
0.961 |
| 228 |
249482_at |
AT5G38980
|
unknown |
0.530 |
0.891 |
| 229 |
247527_at |
AT5G61480
|
PXY, PHLOEM INTERCALATED WITH XYLEM, TDR, TDIF receptor |
0.529 |
-0.153 |
| 230 |
245652_at |
AT4G13960
|
[F-box/RNI-like superfamily protein] |
0.528 |
-0.091 |
| 231 |
266812_at |
AT2G44830
|
[Protein kinase superfamily protein] |
0.528 |
0.391 |
| 232 |
253148_at |
AT4G35620
|
CYCB2;2, Cyclin B2;2 |
0.528 |
0.629 |
| 233 |
261190_at |
AT1G32990
|
PRPL11, plastid ribosomal protein l11 |
0.528 |
0.380 |
| 234 |
259845_at |
AT1G73590
|
ATPIN1, ARABIDOPSIS THALIANA PIN-FORMED 1, PIN1, PIN-FORMED 1 |
0.527 |
0.543 |
| 235 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.526 |
0.334 |
| 236 |
247362_at |
AT5G63140
|
ATPAP29, purple acid phosphatase 29, PAP29, PAP29, purple acid phosphatase 29 |
0.526 |
0.455 |
| 237 |
251962_at |
AT3G53420
|
AtPIP2;1, PIP2, PLASMA MEMBRANE INTRINSIC PROTEIN 2, PIP2;1, PLASMA MEMBRANE INTRINSIC PROTEIN 2;1, PIP2A, plasma membrane intrinsic protein 2A |
0.526 |
0.433 |
| 238 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
0.525 |
0.395 |
| 239 |
251012_at |
AT5G02580
|
unknown |
0.525 |
-0.041 |
| 240 |
262801_at |
AT1G21010
|
unknown |
0.525 |
-0.104 |
| 241 |
254030_at |
AT4G25890
|
[60S acidic ribosomal protein family] |
0.525 |
0.945 |
| 242 |
263264_at |
AT2G38810
|
HTA8, histone H2A 8 |
0.524 |
0.802 |
| 243 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
0.524 |
-0.729 |
| 244 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
0.522 |
0.469 |
| 245 |
256720_at |
AT2G34140
|
[Dof-type zinc finger DNA-binding family protein] |
0.521 |
-0.108 |
| 246 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.520 |
0.215 |
| 247 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
0.520 |
-0.193 |
| 248 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
0.519 |
-0.896 |
| 249 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.519 |
-0.852 |
| 250 |
259151_at |
AT3G10310
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.518 |
0.125 |
| 251 |
245354_at |
AT4G17600
|
LIL3:1 |
0.516 |
0.261 |
| 252 |
249753_at |
AT5G24610
|
unknown |
0.515 |
1.070 |
| 253 |
244962_at |
ATCG01050
|
NDHD |
0.512 |
-0.004 |
| 254 |
253403_at |
AT4G32830
|
AtAUR1, ataurora1, AUR1, ataurora1 |
0.511 |
0.643 |
| 255 |
263209_at |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
0.510 |
1.083 |
| 256 |
265318_at |
AT2G22650
|
[FAD-dependent oxidoreductase family protein] |
0.510 |
0.581 |
| 257 |
258341_at |
AT3G22790
|
NET1A, Networked 1A |
0.510 |
0.039 |
| 258 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
0.509 |
0.947 |
| 259 |
267261_at |
AT2G23120
|
[Late embryogenesis abundant protein, group 6] |
0.507 |
0.087 |
| 260 |
250058_at |
AT5G17870
|
PSRP6, plastid-specific 50S ribosomal protein 6 |
0.506 |
1.045 |
| 261 |
267530_at |
AT2G41890
|
[curculin-like (mannose-binding) lectin family protein / PAN domain-containing protein] |
0.505 |
0.101 |
| 262 |
251935_at |
AT3G54090
|
FLN1, fructokinase-like 1 |
0.505 |
0.416 |
| 263 |
247944_at |
AT5G57100
|
[Nucleotide/sugar transporter family protein] |
0.504 |
-0.124 |
| 264 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
0.503 |
0.598 |
| 265 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.502 |
-0.178 |
| 266 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.502 |
-0.329 |
| 267 |
258532_at |
AT3G06700
|
[Ribosomal L29e protein family] |
0.501 |
1.709 |
| 268 |
249080_at |
AT5G43990
|
SDG18, SET DOMAIN PROTEIN 18, SUVR2 |
0.498 |
-0.296 |
| 269 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
0.497 |
0.759 |
| 270 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
0.496 |
1.143 |
| 271 |
257205_at |
AT3G16520
|
UGT88A1, UDP-glucosyl transferase 88A1 |
0.496 |
1.194 |
| 272 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.495 |
-0.565 |
| 273 |
246005_at |
AT5G08415
|
LIP1, lipoyl synthase 1 |
0.495 |
0.678 |
| 274 |
251067_at |
AT5G01910
|
unknown |
0.495 |
-0.078 |
| 275 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.495 |
0.482 |
| 276 |
262029_at |
AT1G35680
|
RPL21C, chloroplast ribosomal protein L21 |
0.494 |
0.722 |
| 277 |
264061_at |
AT2G27970
|
CKS2, CDK-subunit 2 |
0.493 |
0.258 |
| 278 |
262758_at |
AT1G10780
|
[F-box/RNI-like superfamily protein] |
0.491 |
0.869 |
| 279 |
248691_at |
AT5G48310
|
unknown |
0.490 |
-0.125 |
| 280 |
261549_at |
AT1G63470
|
AHL5, AT-hook motif nuclear localized protein 5 |
0.489 |
0.383 |
| 281 |
259172_at |
AT3G03630
|
CS26, cysteine synthase 26 |
0.488 |
1.260 |
| 282 |
254964_at |
AT4G11080
|
3xHMG-box1, 3xHigh Mobility Group-box1 |
0.488 |
-0.370 |
| 283 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
0.488 |
0.899 |
| 284 |
249778_at |
AT5G24165
|
unknown |
0.487 |
1.417 |
| 285 |
249644_at |
AT5G37010
|
unknown |
0.486 |
1.132 |
| 286 |
263131_at |
AT1G78630
|
emb1473, embryo defective 1473 |
0.485 |
0.845 |
| 287 |
253096_at |
AT4G37330
|
CYP81D4, cytochrome P450, family 81, subfamily D, polypeptide 4 |
0.485 |
0.811 |
| 288 |
261384_at |
AT1G05440
|
[C-8 sterol isomerases] |
0.485 |
0.233 |
| 289 |
244975_at |
ATCG00710
|
PSBH, photosystem II reaction center protein H |
0.485 |
0.617 |
| 290 |
260236_at |
AT1G74470
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
0.484 |
0.584 |
| 291 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.484 |
0.882 |
| 292 |
245912_at |
AT5G19600
|
SULTR3;5, sulfate transporter 3;5 |
0.483 |
-0.029 |
| 293 |
245709_at |
AT5G04320
|
[Shugoshin C terminus] |
0.483 |
0.078 |
| 294 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
0.482 |
1.075 |
| 295 |
260967_at |
AT1G12230
|
[Aldolase superfamily protein] |
0.480 |
0.986 |
| 296 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.480 |
0.673 |
| 297 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
0.479 |
0.239 |
| 298 |
249635_at |
AT5G36870
|
ATGSL09, glucan synthase-like 9, atgsl9, gsl09, GSL09, glucan synthase-like 9 |
0.478 |
0.325 |
| 299 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
0.478 |
0.035 |
| 300 |
265247_at |
AT2G43030
|
[Ribosomal protein L3 family protein] |
0.478 |
0.510 |
| 301 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
-1.710 |
-1.733 |
| 302 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-1.559 |
-0.888 |
| 303 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-1.404 |
-0.334 |
| 304 |
259385_at |
AT1G13470
|
unknown |
-1.400 |
-0.671 |
| 305 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
-1.381 |
-0.545 |
| 306 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-1.319 |
0.127 |
| 307 |
255016_at |
AT4G10120
|
ATSPS4F |
-1.312 |
-0.351 |
| 308 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
-1.283 |
-0.726 |
| 309 |
249134_at |
AT5G43150
|
unknown |
-1.232 |
0.133 |
| 310 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-1.210 |
-0.131 |
| 311 |
246001_at |
AT5G20790
|
unknown |
-1.209 |
-0.020 |
| 312 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-1.209 |
-0.461 |
| 313 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.178 |
-0.768 |
| 314 |
249465_at |
AT5G39720
|
AIG2L, avirulence induced gene 2 like protein |
-1.136 |
-0.741 |
| 315 |
254373_at |
AT4G21730
|
[expressed protein] |
-1.096 |
-0.495 |
| 316 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-1.087 |
-1.009 |
| 317 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
-1.075 |
-0.888 |
| 318 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-1.024 |
-0.232 |
| 319 |
251612_at |
AT3G57950
|
unknown |
-1.023 |
-0.578 |
| 320 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.995 |
0.179 |
| 321 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-0.995 |
-0.841 |
| 322 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-0.988 |
-0.251 |
| 323 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.985 |
-0.323 |
| 324 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.985 |
-0.248 |
| 325 |
249009_at |
AT5G44610
|
MAP18, microtubule-associated protein 18, PCAP2, PLASMA MEMBRANE ASSOCIATED CA2+-BINDING PROTEIN-2 |
-0.969 |
0.121 |
| 326 |
252730_at |
AT3G43110
|
unknown |
-0.962 |
-0.207 |
| 327 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
-0.961 |
0.440 |
| 328 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.956 |
-0.446 |
| 329 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.954 |
-1.026 |
| 330 |
253087_at |
AT4G36350
|
ATPAP25, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 25, PAP25, purple acid phosphatase 25 |
-0.944 |
-0.781 |
| 331 |
249893_at |
AT5G22555
|
unknown |
-0.943 |
-0.581 |
| 332 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.935 |
-0.617 |
| 333 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.922 |
-0.835 |
| 334 |
260040_at |
AT1G68765
|
IDA, INFLORESCENCE DEFICIENT IN ABSCISSION |
-0.921 |
-0.002 |
| 335 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.919 |
-0.178 |
| 336 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.905 |
-0.705 |
| 337 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.893 |
-0.063 |
| 338 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.892 |
-0.439 |
| 339 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.892 |
0.026 |
| 340 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.879 |
-0.971 |
| 341 |
249150_at |
AT5G43340
|
PHT1;6, phosphate transporter 1;6, PHT6, PHOSPHATE TRANSPORTER 6 |
-0.864 |
-0.974 |
| 342 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.854 |
-0.424 |
| 343 |
260415_at |
AT1G69790
|
[Protein kinase superfamily protein] |
-0.853 |
0.066 |
| 344 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.851 |
-0.026 |
| 345 |
260239_at |
AT1G74360
|
[Leucine-rich repeat protein kinase family protein] |
-0.847 |
-0.452 |
| 346 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
-0.846 |
-0.564 |
| 347 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
-0.846 |
-0.996 |
| 348 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.842 |
-0.148 |
| 349 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.837 |
0.612 |
| 350 |
251315_at |
AT3G61410
|
unknown |
-0.837 |
-0.916 |
| 351 |
266376_at |
AT2G14620
|
XTH10, xyloglucan endotransglucosylase/hydrolase 10 |
-0.836 |
-0.345 |
| 352 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.832 |
-0.302 |
| 353 |
259479_at |
AT1G19020
|
unknown |
-0.830 |
-0.031 |
| 354 |
252539_at |
AT3G45730
|
unknown |
-0.824 |
-1.496 |
| 355 |
251792_at |
AT3G55550
|
LecRK-S.4, L-type lectin receptor kinase S.4 |
-0.821 |
-1.130 |
| 356 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.820 |
-0.620 |
| 357 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
-0.817 |
0.194 |
| 358 |
256759_at |
AT3G25640
|
unknown |
-0.815 |
0.150 |
| 359 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.809 |
-0.727 |
| 360 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.805 |
-0.740 |
| 361 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-0.789 |
-0.708 |
| 362 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.789 |
-0.779 |
| 363 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
-0.784 |
-0.499 |
| 364 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
-0.780 |
-1.168 |
| 365 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
-0.774 |
-0.637 |
| 366 |
246076_at |
AT5G20280
|
ATSPS1F, sucrose phosphate synthase 1F, SPS1F, sucrose phosphate synthase 1F, SPSA1, sucrose-phosphate synthase A1 |
-0.772 |
0.497 |
| 367 |
264252_at |
AT1G09180
|
ATSAR1, SECRETION-ASSOCIATED RAS 1, ATSARA1A, secretion-associated RAS super family 1, SARA1A, secretion-associated RAS super family 1 |
-0.771 |
-0.175 |
| 368 |
266757_at |
AT2G46940
|
unknown |
-0.770 |
0.216 |
| 369 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.758 |
0.101 |
| 370 |
245563_at |
AT4G14580
|
CIPK4, CBL-interacting protein kinase 4, SnRK3.3, SNF1-RELATED PROTEIN KINASE 3.3 |
-0.757 |
0.100 |
| 371 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
-0.742 |
-0.252 |
| 372 |
248889_at |
AT5G46230
|
unknown |
-0.741 |
-0.222 |
| 373 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.737 |
0.372 |
| 374 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
-0.733 |
-0.199 |
| 375 |
254574_at |
AT4G19430
|
unknown |
-0.733 |
-0.487 |
| 376 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.732 |
-0.161 |
| 377 |
252161_at |
AT3G50580
|
[LOCATED IN: endomembrane system] |
-0.726 |
-0.946 |
| 378 |
267279_at |
AT2G19460
|
unknown |
-0.724 |
-0.314 |
| 379 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.723 |
0.067 |
| 380 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.721 |
-0.901 |
| 381 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
-0.719 |
-0.946 |
| 382 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
-0.719 |
-0.655 |
| 383 |
258845_at |
AT3G03150
|
unknown |
-0.716 |
0.456 |
| 384 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.716 |
0.069 |
| 385 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.716 |
-0.912 |
| 386 |
264305_at |
AT1G78815
|
LSH7, LIGHT SENSITIVE HYPOCOTYLS 7 |
-0.712 |
-0.339 |
| 387 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
-0.711 |
-0.749 |
| 388 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
-0.708 |
-0.265 |
| 389 |
260201_at |
AT1G67600
|
[Acid phosphatase/vanadium-dependent haloperoxidase-related protein] |
-0.706 |
0.142 |
| 390 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-0.703 |
0.319 |
| 391 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.702 |
0.164 |
| 392 |
257421_at |
AT1G12030
|
unknown |
-0.701 |
-0.278 |
| 393 |
266017_at |
AT2G18690
|
unknown |
-0.698 |
-0.163 |
| 394 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
-0.696 |
-0.195 |
| 395 |
254111_at |
AT4G24890
|
ATPAP24, ARABIDOPSIS THALIANA PURPLE ACID PHOSPHATASE 24, PAP24, purple acid phosphatase 24 |
-0.694 |
-1.021 |
| 396 |
257810_at |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
-0.693 |
-0.427 |
| 397 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.686 |
-0.444 |
| 398 |
255950_at |
AT1G22110
|
[structural constituent of ribosome] |
-0.685 |
-0.570 |
| 399 |
261056_at |
AT1G01360
|
PYL9, PYRABACTIN RESISTANCE 1-LIKE 9, RCAR1, regulatory component of ABA receptor 1 |
-0.683 |
0.186 |
| 400 |
250881_at |
AT5G04080
|
unknown |
-0.680 |
-1.177 |
| 401 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
-0.680 |
-0.859 |
| 402 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
-0.675 |
-0.732 |
| 403 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
-0.675 |
-0.966 |
| 404 |
253228_at |
AT4G34630
|
unknown |
-0.674 |
0.247 |
| 405 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.674 |
0.109 |
| 406 |
250618_at |
AT5G07220
|
ATBAG3, BCL-2-associated athanogene 3, BAG3, BCL-2-associated athanogene 3 |
-0.673 |
-0.280 |
| 407 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.671 |
-0.669 |
| 408 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
-0.670 |
0.080 |
| 409 |
255587_at |
AT4G01480
|
AtPPa5, pyrophosphorylase 5, PPa5, pyrophosphorylase 5 |
-0.669 |
0.558 |
| 410 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
-0.668 |
-1.493 |
| 411 |
254571_at |
AT4G19370
|
unknown |
-0.668 |
-0.756 |
| 412 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
-0.666 |
-0.701 |
| 413 |
247051_at |
AT5G66670
|
unknown |
-0.663 |
0.113 |
| 414 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.662 |
-0.982 |
| 415 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.662 |
0.723 |
| 416 |
258078_at |
AT3G25870
|
unknown |
-0.660 |
-0.049 |
| 417 |
253014_at |
AT4G37940
|
AGL21, AGAMOUS-like 21 |
-0.659 |
-0.362 |
| 418 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.653 |
-0.357 |
| 419 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
-0.652 |
-0.452 |
| 420 |
265387_at |
AT2G20670
|
unknown |
-0.651 |
0.246 |
| 421 |
259852_at |
AT1G72280
|
AERO1, endoplasmic reticulum oxidoreductins 1, ERO1, endoplasmic reticulum oxidoreductins 1 |
-0.647 |
-1.040 |
| 422 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.647 |
-0.373 |
| 423 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.647 |
-0.035 |
| 424 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.645 |
-0.587 |
| 425 |
262314_at |
AT1G70810
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.644 |
-0.561 |
| 426 |
260403_at |
AT1G69810
|
ATWRKY36, WRKY36, WRKY DNA-binding protein 36 |
-0.643 |
-0.926 |
| 427 |
245028_at |
AT2G26570
|
WEB1, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1 |
-0.641 |
0.112 |
| 428 |
251761_at |
AT3G55700
|
[UDP-Glycosyltransferase superfamily protein] |
-0.641 |
-0.514 |
| 429 |
245882_at |
AT5G09470
|
DIC3, dicarboxylate carrier 3 |
-0.639 |
-0.113 |
| 430 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.639 |
-1.379 |
| 431 |
253963_at |
AT4G26470
|
[Calcium-binding EF-hand family protein] |
-0.638 |
-0.837 |
| 432 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
-0.637 |
-0.172 |
| 433 |
258362_at |
AT3G14280
|
unknown |
-0.636 |
-0.824 |
| 434 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.634 |
0.452 |
| 435 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.631 |
-0.665 |
| 436 |
259887_at |
AT1G76360
|
[Protein kinase superfamily protein] |
-0.629 |
-0.837 |
| 437 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.629 |
-0.763 |
| 438 |
260002_at |
AT1G67940
|
ABCI17, ATP-binding cassette I17, ATNAP3, non-intrinsic ABC protein 3, AtSTAR1, NAP3, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 3, NAP3, non-intrinsic ABC protein 3 |
-0.629 |
-0.014 |
| 439 |
256014_at |
AT1G19200
|
unknown |
-0.629 |
-0.357 |
| 440 |
262350_at |
AT2G48150
|
ATGPX4, glutathione peroxidase 4, GPX4, glutathione peroxidase 4 |
-0.628 |
-1.755 |
| 441 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.626 |
-0.037 |
| 442 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.623 |
-0.068 |
| 443 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.623 |
-0.780 |
| 444 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.623 |
1.082 |
| 445 |
255608_at |
AT4G01140
|
unknown |
-0.620 |
0.083 |
| 446 |
252400_at |
AT3G48020
|
unknown |
-0.618 |
-0.241 |
| 447 |
262228_at |
AT1G68690
|
AtPERK9, proline-rich extensin-like receptor kinase 9, PERK9, proline-rich extensin-like receptor kinase 9 |
-0.617 |
-0.055 |
| 448 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
-0.616 |
0.837 |
| 449 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.616 |
-0.539 |
| 450 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
-0.613 |
-0.726 |
| 451 |
256256_at |
AT3G11230
|
[Yippee family putative zinc-binding protein] |
-0.612 |
-0.267 |
| 452 |
258203_at |
AT3G13950
|
unknown |
-0.610 |
-0.269 |
| 453 |
255502_at |
AT4G02410
|
AtLPK1, LecRK-IV.3, L-type lectin receptor kinase IV.3, LPK1, lectin-like protein kinase 1 |
-0.609 |
-0.779 |
| 454 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.609 |
-0.457 |
| 455 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
-0.607 |
-1.123 |
| 456 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.604 |
-1.382 |
| 457 |
254097_at |
AT4G25160
|
[U-box domain-containing protein kinase family protein] |
-0.597 |
-0.768 |
| 458 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
-0.597 |
0.091 |
| 459 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
-0.595 |
0.439 |
| 460 |
250241_at |
AT5G13600
|
[Phototropic-responsive NPH3 family protein] |
-0.594 |
-0.256 |
| 461 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
-0.594 |
0.166 |
| 462 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
-0.593 |
0.138 |
| 463 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.592 |
-0.805 |
| 464 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.591 |
-0.878 |
| 465 |
245629_at |
AT1G56580
|
SVB, SMALLER WITH VARIABLE BRANCHES |
-0.588 |
0.690 |
| 466 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.585 |
-0.770 |
| 467 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
-0.583 |
-0.320 |
| 468 |
261900_at |
AT1G80940
|
unknown |
-0.583 |
-0.074 |
| 469 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.583 |
-0.309 |
| 470 |
263987_at |
AT2G42690
|
[alpha/beta-Hydrolases superfamily protein] |
-0.583 |
0.991 |
| 471 |
257846_at |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-0.581 |
-0.392 |
| 472 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.581 |
-0.518 |
| 473 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.580 |
-0.830 |
| 474 |
263378_at |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
-0.578 |
-0.832 |
| 475 |
258786_at |
AT3G11820
|
AT-SYR1, ATSYP121, ATSYR1, SYNTAXIN RELATED PROTEIN 1, PEN1, PENETRATION1, SYP121, syntaxin of plants 121, SYR1, SYNTAXIN RELATED PROTEIN 1 |
-0.574 |
0.070 |
| 476 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.573 |
-0.359 |
| 477 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
-0.573 |
1.280 |
| 478 |
259921_at |
AT1G72540
|
[Protein kinase superfamily protein] |
-0.573 |
-0.748 |
| 479 |
254524_at |
AT4G20000
|
[VQ motif-containing protein] |
-0.572 |
-0.404 |
| 480 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.569 |
-0.869 |
| 481 |
253454_at |
AT4G31875
|
unknown |
-0.567 |
-0.121 |
| 482 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.567 |
-0.587 |
| 483 |
252253_at |
AT3G49300
|
[proline-rich family protein] |
-0.566 |
-0.398 |
| 484 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-0.566 |
-0.401 |
| 485 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.565 |
-0.341 |
| 486 |
255627_at |
AT4G00955
|
unknown |
-0.562 |
0.135 |
| 487 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
-0.561 |
0.338 |
| 488 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.561 |
-0.230 |
| 489 |
247597_at |
AT5G60860
|
AtRABA1f, RAB GTPase homolog A1F, RABA1f, RAB GTPase homolog A1F |
-0.561 |
-0.181 |
| 490 |
253264_at |
AT4G33950
|
ATOST1, OPEN STOMATA 1, OST1, OPEN STOMATA 1, P44, SNRK2-6, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-6, SNRK2.6, SNF1-RELATED PROTEIN KINASE 2.6, SRK2E |
-0.560 |
0.935 |
| 491 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.560 |
-0.448 |
| 492 |
257226_at |
AT3G27880
|
unknown |
-0.560 |
0.390 |
| 493 |
253626_at |
AT4G30640
|
[RNI-like superfamily protein] |
-0.559 |
-0.985 |
| 494 |
246200_at |
AT4G37240
|
unknown |
-0.558 |
1.011 |
| 495 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.557 |
0.333 |
| 496 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.557 |
-0.959 |
| 497 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.555 |
-0.723 |
| 498 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.555 |
0.310 |
| 499 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
-0.554 |
-1.205 |
| 500 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
-0.553 |
0.155 |
| 501 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
-0.552 |
0.457 |
| 502 |
266685_at |
AT2G19710
|
[Regulator of Vps4 activity in the MVB pathway protein] |
-0.552 |
-0.041 |
| 503 |
266911_at |
AT2G45910
|
[U-box domain-containing protein kinase family protein] |
-0.551 |
-0.254 |
| 504 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
-0.551 |
-0.358 |
| 505 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.551 |
-0.532 |
| 506 |
261984_at |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-0.550 |
-1.134 |
| 507 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
-0.549 |
0.270 |
| 508 |
258023_at |
AT3G19450
|
ATCAD4, CAD, CAD-C, CAD4, CINNAMYL ALCOHOL DEHYDROGENASE 4 |
-0.549 |
0.765 |
| 509 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
-0.549 |
-1.119 |
| 510 |
266380_at |
AT2G14680
|
MEE13, maternal effect embryo arrest 13 |
-0.549 |
0.542 |
| 511 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.548 |
0.033 |
| 512 |
260896_at |
AT1G29310
|
[SecY protein transport family protein] |
-0.548 |
0.360 |
| 513 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
-0.547 |
-0.775 |
| 514 |
246012_at |
AT5G10650
|
[RING/U-box superfamily protein] |
-0.546 |
-0.535 |
| 515 |
259334_at |
AT3G03790
|
[ankyrin repeat family protein / regulator of chromosome condensation (RCC1) family protein] |
-0.546 |
0.779 |
| 516 |
248366_at |
AT5G52510
|
SCL8, SCARECROW-like 8 |
-0.546 |
-0.024 |
| 517 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
-0.545 |
0.437 |
| 518 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.543 |
-0.463 |
| 519 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
-0.542 |
-0.094 |
| 520 |
245756_at |
AT1G35190
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.541 |
-0.340 |
| 521 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
-0.539 |
-0.223 |
| 522 |
249476_at |
AT5G38910
|
[RmlC-like cupins superfamily protein] |
-0.538 |
-0.559 |
| 523 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
-0.537 |
-0.382 |
| 524 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
-0.537 |
-0.432 |
| 525 |
261120_at |
AT1G75410
|
BLH3, BEL1-like homeodomain 3 |
-0.536 |
-0.452 |
| 526 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.535 |
-0.105 |
| 527 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.535 |
-0.346 |
| 528 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.534 |
-0.761 |
| 529 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.534 |
-0.760 |
| 530 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.534 |
-0.102 |
| 531 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.531 |
-0.753 |
| 532 |
255561_at |
AT4G02050
|
STP7, sugar transporter protein 7 |
-0.531 |
0.560 |
| 533 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.531 |
-0.310 |
| 534 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
-0.530 |
-1.182 |
| 535 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.530 |
-0.624 |
| 536 |
252006_at |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
-0.529 |
-0.642 |
| 537 |
262072_at |
AT1G59590
|
ZCF37 |
-0.529 |
0.432 |
| 538 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.528 |
-0.003 |
| 539 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
-0.526 |
-0.244 |
| 540 |
249550_at |
AT5G38210
|
[Protein kinase family protein] |
-0.526 |
0.391 |
| 541 |
247105_at |
AT5G66630
|
DAR5, DA1-related protein 5 |
-0.525 |
-0.249 |
| 542 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
-0.525 |
0.527 |
| 543 |
253676_at |
AT4G29570
|
[Cytidine/deoxycytidylate deaminase family protein] |
-0.523 |
-0.633 |
| 544 |
259015_at |
AT3G07350
|
unknown |
-0.523 |
-0.313 |
| 545 |
267209_at |
AT2G30930
|
unknown |
-0.522 |
-0.540 |
| 546 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.521 |
-0.256 |
| 547 |
252345_at |
AT3G48640
|
unknown |
-0.521 |
-0.831 |
| 548 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
-0.519 |
0.563 |
| 549 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.517 |
0.461 |
| 550 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
-0.516 |
0.225 |
| 551 |
253774_at |
AT4G28530
|
anac074, NAC domain containing protein 74, NAC074, NAC domain containing protein 74 |
-0.516 |
-0.201 |
| 552 |
249705_at |
AT5G35580
|
[Protein kinase superfamily protein] |
-0.513 |
0.216 |
| 553 |
255756_at |
AT1G19940
|
AtGH9B5, glycosyl hydrolase 9B5, GH9B5, glycosyl hydrolase 9B5 |
-0.513 |
-1.163 |
| 554 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
-0.513 |
-0.499 |
| 555 |
245621_at |
AT4G14070
|
AAE15, acyl-activating enzyme 15 |
-0.512 |
0.147 |
| 556 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
-0.512 |
1.120 |
| 557 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
-0.511 |
0.422 |
| 558 |
250345_at |
AT5G11940
|
[Subtilase family protein] |
-0.510 |
-0.595 |
| 559 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
-0.510 |
0.299 |
| 560 |
254921_at |
AT4G11300
|
unknown |
-0.509 |
-0.051 |
| 561 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
-0.509 |
0.526 |
| 562 |
253257_at |
AT4G34390
|
XLG2, extra-large GTP-binding protein 2 |
-0.508 |
-0.355 |
| 563 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.508 |
0.237 |
| 564 |
263419_at |
AT2G17220
|
Kin3, kinase 3 |
-0.507 |
-0.125 |
| 565 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.507 |
-0.140 |
| 566 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
-0.504 |
-0.355 |
| 567 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.503 |
-0.583 |
| 568 |
251479_at |
AT3G59700
|
ATHLECRK, lectin-receptor kinase, HLECRK, lectin-receptor kinase, LecRK-V.5, L-type lectin receptor kinase V.5, LecRK-V.5, LEGUME-LIKE LECTIN RECEPTOR KINASE-V.5, LECRK1, LECTIN-RECEPTOR KINASE 1 |
-0.503 |
-1.046 |
| 569 |
266302_at |
AT2G27050
|
AtEIL1, EIL1, ETHYLENE-INSENSITIVE3-like 1 |
-0.502 |
-0.446 |
| 570 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.501 |
-0.914 |
| 571 |
265470_at |
AT2G37150
|
[RING/U-box superfamily protein] |
-0.501 |
-0.313 |
| 572 |
266037_at |
AT2G05940
|
RIPK, RPM1-induced protein kinase |
-0.497 |
0.989 |
| 573 |
264145_at |
AT1G79310
|
AtMC7, metacaspase 7, AtMCP2a, metacaspase 2a, MC7, metacaspase 7, MCP2a, metacaspase 2a |
-0.496 |
0.149 |
| 574 |
249004_at |
AT5G44570
|
unknown |
-0.493 |
-1.015 |
| 575 |
259750_at |
AT1G71130
|
AtERF070, CRF8, cytokinin response factor 8, ERF070, ethylene response factor 70 |
-0.492 |
0.722 |
| 576 |
247594_at |
AT5G60800
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.492 |
0.216 |
| 577 |
245566_at |
AT4G14610
|
[pseudogene] |
-0.491 |
-0.118 |
| 578 |
262220_at |
AT1G74740
|
ATCPK30, CDPK1A, CALCIUM-DEPENDENT PROTEIN KINASE 1A, CPK30, calcium-dependent protein kinase 30 |
-0.491 |
-0.931 |
| 579 |
266330_at |
AT2G01530
|
MLP329, MLP-like protein 329, ZCE2, (Zusammen-CA)-enhanced 2 |
-0.491 |
0.216 |
| 580 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.490 |
-0.298 |
| 581 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.490 |
0.490 |
| 582 |
254796_at |
AT4G13000
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.489 |
-0.621 |
| 583 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.487 |
-0.643 |
| 584 |
255546_at |
AT4G01910
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.486 |
0.030 |
| 585 |
260843_at |
AT1G29060
|
[Target SNARE coiled-coil domain protein] |
-0.485 |
-0.429 |
| 586 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
-0.485 |
0.230 |
| 587 |
263920_at |
AT2G36410
|
unknown |
-0.485 |
0.389 |
| 588 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
-0.484 |
-0.842 |
| 589 |
263545_at |
AT2G21560
|
unknown |
-0.484 |
0.093 |
| 590 |
255953_at |
AT1G22070
|
TGA3, TGA1A-related gene 3 |
-0.484 |
-0.143 |
| 591 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.483 |
-0.450 |
| 592 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.482 |
0.473 |
| 593 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
-0.482 |
-0.207 |
| 594 |
247045_at |
AT5G66930
|
unknown |
-0.482 |
0.760 |
| 595 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.482 |
-0.569 |
| 596 |
264717_at |
AT1G70140
|
ATFH8, formin 8, FH8, formin 8 |
-0.482 |
0.004 |
| 597 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.481 |
0.890 |
| 598 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.481 |
-0.128 |
| 599 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
-0.479 |
-0.179 |
| 600 |
258682_at |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
-0.479 |
-0.405 |