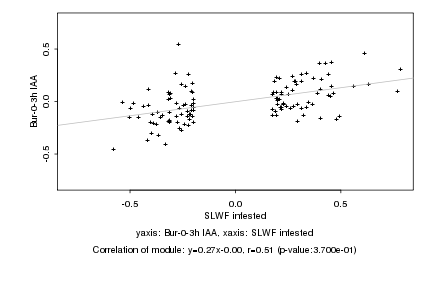
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
SLWF infested |
Log2 signal ratio
Bur-0-3h IAA |
| 1 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.781 |
0.311 |
| 2 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.768 |
0.099 |
| 3 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.629 |
0.166 |
| 4 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.611 |
0.460 |
| 5 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.558 |
0.152 |
| 6 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
0.492 |
-0.134 |
| 7 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.475 |
-0.165 |
| 8 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.465 |
0.078 |
| 9 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.454 |
0.376 |
| 10 |
257154_at |
AT3G27210
|
unknown |
0.453 |
0.152 |
| 11 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.447 |
0.057 |
| 12 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.439 |
0.259 |
| 13 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.439 |
0.066 |
| 14 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
0.423 |
0.366 |
| 15 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.408 |
0.216 |
| 16 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
0.402 |
-0.159 |
| 17 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.399 |
0.118 |
| 18 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.395 |
0.370 |
| 19 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.387 |
0.078 |
| 20 |
256833_at |
AT3G22910
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.366 |
0.220 |
| 21 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.362 |
-0.022 |
| 22 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.346 |
-0.001 |
| 23 |
254243_at |
AT4G23210
|
CRK13, cysteine-rich RLK (RECEPTOR-like protein kinase) 13 |
0.336 |
0.268 |
| 24 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.333 |
-0.053 |
| 25 |
265658_at |
AT2G13810
|
ALD1, AGD2-like defense response protein 1, AtALD1, EDTS5, eds two suppressor 5 |
0.320 |
-0.126 |
| 26 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.313 |
0.196 |
| 27 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.311 |
0.263 |
| 28 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.309 |
-0.057 |
| 29 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.293 |
-0.181 |
| 30 |
246309_at |
AT3G51790
|
AtCCME, A. thaliana homolog of E. coli CcmE, ATG1, transmembrane protein G1P-related 1, G1, transmembrane protein G1P-related 1 |
0.290 |
-0.025 |
| 31 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
0.289 |
0.163 |
| 32 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.281 |
0.200 |
| 33 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.279 |
0.199 |
| 34 |
258813_at |
AT3G04060
|
anac046, NAC domain containing protein 46, NAC046, NAC domain containing protein 46 |
0.275 |
-0.046 |
| 35 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.267 |
0.242 |
| 36 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.266 |
0.111 |
| 37 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.258 |
-0.060 |
| 38 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.251 |
0.071 |
| 39 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.240 |
0.142 |
| 40 |
260771_at |
AT1G49160
|
WNK7 |
0.238 |
-0.042 |
| 41 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
0.227 |
-0.021 |
| 42 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.227 |
-0.020 |
| 43 |
251063_at |
AT5G01850
|
[Protein kinase superfamily protein] |
0.226 |
-0.014 |
| 44 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.217 |
0.071 |
| 45 |
250776_at |
AT5G05320
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.216 |
0.095 |
| 46 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.216 |
-0.075 |
| 47 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.215 |
-0.055 |
| 48 |
266464_at |
AT2G47800
|
ABCC4, ATP-binding cassette C4, ATMRP4, multidrug resistance-associated protein 4, EST3, MRP4, multidrug resistance-associated protein 4 |
0.209 |
-0.051 |
| 49 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.207 |
0.228 |
| 50 |
261934_at |
AT1G22400
|
ATUGT85A1, ARABIDOPSIS THALIANA UDP-GLUCOSYL TRANSFERASE 85A1, UGT85A1 |
0.205 |
0.025 |
| 51 |
259403_at |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
0.199 |
0.018 |
| 52 |
249112_at |
AT5G43780
|
APS4 |
0.198 |
-0.026 |
| 53 |
259661_at |
AT1G55265
|
unknown |
0.197 |
0.038 |
| 54 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.194 |
0.031 |
| 55 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
0.193 |
0.092 |
| 56 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.191 |
0.236 |
| 57 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.190 |
-0.126 |
| 58 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
0.188 |
-0.086 |
| 59 |
259137_at |
AT3G10300
|
[Calcium-binding EF-hand family protein] |
0.184 |
0.198 |
| 60 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.179 |
0.091 |
| 61 |
256433_at |
AT3G10985
|
ATWI-12, ARABIDOPSIS THALIANA WOUND-INDUCED PROTEIN 12, SAG20, senescence associated gene 20, WI12, WOUND-INDUCED PROTEIN 12 |
0.174 |
0.069 |
| 62 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.174 |
-0.072 |
| 63 |
262281_at |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
0.173 |
-0.124 |
| 64 |
247814_at |
AT5G58310
|
ATMES18, ARABIDOPSIS THALIANA METHYL ESTERASE 18, MES18, methyl esterase 18 |
-0.583 |
-0.449 |
| 65 |
248509_at |
AT5G50335
|
unknown |
-0.540 |
-0.004 |
| 66 |
248975_at |
AT5G45040
|
CYTC6A, cytochrome c6A |
-0.504 |
-0.143 |
| 67 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.498 |
-0.061 |
| 68 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
-0.486 |
-0.018 |
| 69 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.461 |
-0.148 |
| 70 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.439 |
-0.038 |
| 71 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.421 |
-0.367 |
| 72 |
266613_at |
AT2G14900
|
[Gibberellin-regulated family protein] |
-0.417 |
-0.034 |
| 73 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.413 |
0.122 |
| 74 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.406 |
-0.196 |
| 75 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.401 |
-0.300 |
| 76 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.398 |
-0.122 |
| 77 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.391 |
-0.209 |
| 78 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.376 |
-0.217 |
| 79 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.373 |
-0.103 |
| 80 |
249472_at |
AT5G39210
|
CRR7, CHLORORESPIRATORY REDUCTION 7 |
-0.368 |
-0.319 |
| 81 |
261931_at |
AT1G22430
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.356 |
-0.152 |
| 82 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-0.350 |
-0.127 |
| 83 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.336 |
-0.406 |
| 84 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.322 |
-0.182 |
| 85 |
260112_at |
AT1G63310
|
unknown |
-0.319 |
0.028 |
| 86 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.319 |
0.090 |
| 87 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.317 |
0.071 |
| 88 |
248186_at |
AT5G53880
|
unknown |
-0.317 |
-0.100 |
| 89 |
266285_at |
AT2G29180
|
unknown |
-0.315 |
-0.195 |
| 90 |
247977_at |
AT5G56850
|
unknown |
-0.315 |
-0.183 |
| 91 |
261638_at |
AT1G49975
|
[INVOLVED IN: photosynthesis] |
-0.314 |
-0.172 |
| 92 |
247358_at |
AT5G63580
|
ATFLS2, FLS2, flavonol synthase 2 |
-0.312 |
0.081 |
| 93 |
258796_at |
AT3G04630
|
WDL1, WVD2-like 1 |
-0.311 |
0.033 |
| 94 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.288 |
0.267 |
| 95 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.280 |
-0.010 |
| 96 |
251008_at |
AT5G02710
|
unknown |
-0.280 |
-0.137 |
| 97 |
256698_at |
AT3G20680
|
unknown |
-0.276 |
-0.197 |
| 98 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.272 |
0.547 |
| 99 |
261742_at |
AT1G08390
|
unknown |
-0.270 |
-0.063 |
| 100 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.266 |
-0.254 |
| 101 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.260 |
-0.271 |
| 102 |
265116_at |
AT1G62480
|
[Vacuolar calcium-binding protein-related] |
-0.258 |
-0.119 |
| 103 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.257 |
0.168 |
| 104 |
253589_at |
AT4G30825
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.251 |
-0.036 |
| 105 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
-0.246 |
-0.211 |
| 106 |
249887_at |
AT5G22310
|
unknown |
-0.240 |
-0.027 |
| 107 |
248789_at |
AT5G47440
|
unknown |
-0.238 |
0.147 |
| 108 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.230 |
-0.140 |
| 109 |
250503_at |
AT5G09820
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-0.229 |
-0.089 |
| 110 |
261327_at |
AT1G44830
|
[Integrase-type DNA-binding superfamily protein] |
-0.226 |
0.265 |
| 111 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.224 |
-0.221 |
| 112 |
260260_at |
AT1G68540
|
CCRL6, cinnamoyl coA reductase-like 6, TKPR2, tetraketide alpha-pyrone reductase 2 |
-0.221 |
-0.122 |
| 113 |
266813_at |
AT2G44920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.218 |
-0.163 |
| 114 |
255572_at |
AT4G01050
|
TROL, thylakoid rhodanese-like |
-0.214 |
-0.116 |
| 115 |
263678_at |
AT1G04420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.213 |
-0.083 |
| 116 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
-0.209 |
0.097 |
| 117 |
248911_at |
AT5G45830
|
ATDOG1, DOG1, DELAY OF GERMINATION 1, GAAS5, germination ability after storage 5, GSQ5, GLUCOSE SENSING QTL 5 |
-0.209 |
-0.031 |
| 118 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.208 |
0.087 |
| 119 |
250201_at |
AT5G14230
|
unknown |
-0.204 |
0.180 |
| 120 |
262854_at |
AT1G20870
|
[HSP20-like chaperones superfamily protein] |
-0.204 |
-0.139 |
| 121 |
245396_at |
AT4G14870
|
SECE1 |
-0.202 |
-0.084 |
| 122 |
264523_at |
AT1G10030
|
ERG28, homolog of yeast ergosterol28 |
-0.201 |
0.021 |
| 123 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
-0.201 |
-0.016 |
| 124 |
248285_at |
AT5G52960
|
unknown |
-0.201 |
-0.055 |
| 125 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.199 |
-0.191 |