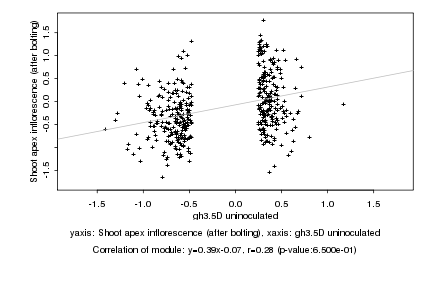
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gh3.5D uninoculated |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
1.166 |
-0.062 |
| 2 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.800 |
-0.774 |
| 3 |
246516_at |
AT5G15740
|
[O-fucosyltransferase family protein] |
0.713 |
0.763 |
| 4 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.711 |
0.114 |
| 5 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.680 |
-0.202 |
| 6 |
259001_at |
AT3G01960
|
unknown |
0.666 |
-0.260 |
| 7 |
249469_at |
AT5G39320
|
UDG4, UDP-glucose dehydrogenase 4 |
0.658 |
0.928 |
| 8 |
245692_at |
AT5G04150
|
BHLH101 |
0.645 |
-0.556 |
| 9 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.642 |
0.296 |
| 10 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.630 |
-0.375 |
| 11 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.627 |
-0.855 |
| 12 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
0.615 |
-0.620 |
| 13 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.605 |
-1.073 |
| 14 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
0.597 |
-0.243 |
| 15 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.573 |
-1.170 |
| 16 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.554 |
-0.682 |
| 17 |
259432_at |
AT1G01520
|
ASG4, ALTERED SEED GERMINATION 4 |
0.547 |
-0.316 |
| 18 |
256021_at |
AT1G58270
|
ZW9 |
0.544 |
-0.195 |
| 19 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.540 |
0.906 |
| 20 |
248789_at |
AT5G47440
|
unknown |
0.532 |
-0.442 |
| 21 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.530 |
0.002 |
| 22 |
259760_at |
AT1G77580
|
unknown |
0.525 |
-0.247 |
| 23 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.515 |
1.122 |
| 24 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
0.514 |
0.314 |
| 25 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.513 |
-0.199 |
| 26 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.506 |
-0.106 |
| 27 |
252215_at |
AT3G50240
|
KICP-02 |
0.505 |
-0.361 |
| 28 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
0.493 |
0.504 |
| 29 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.491 |
-0.958 |
| 30 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.487 |
0.622 |
| 31 |
257759_at |
AT3G23070
|
ATCFM3A, CFM3A, CRM family member 3A |
0.482 |
-0.148 |
| 32 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.481 |
0.718 |
| 33 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.459 |
-0.497 |
| 34 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
0.457 |
0.915 |
| 35 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.454 |
0.152 |
| 36 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
0.454 |
0.754 |
| 37 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
0.449 |
0.054 |
| 38 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
0.449 |
0.717 |
| 39 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.449 |
-0.189 |
| 40 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
0.448 |
-0.045 |
| 41 |
257798_at |
AT3G15950
|
NAI2 |
0.446 |
0.318 |
| 42 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.446 |
-0.410 |
| 43 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
0.445 |
-0.355 |
| 44 |
254122_at |
AT4G24510
|
CER2, ECERIFERUM 2, VC-2, VC2 |
0.445 |
1.119 |
| 45 |
245616_at |
AT4G14480
|
[Protein kinase superfamily protein] |
0.441 |
-0.596 |
| 46 |
261150_at |
AT1G19640
|
JMT, jasmonic acid carboxyl methyltransferase |
0.435 |
-0.499 |
| 47 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
0.435 |
0.248 |
| 48 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.431 |
-0.452 |
| 49 |
256675_at |
AT3G52170
|
[DNA binding] |
0.431 |
-0.132 |
| 50 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
0.427 |
0.168 |
| 51 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
0.424 |
0.345 |
| 52 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
0.423 |
-0.242 |
| 53 |
245404_at |
AT4G17610
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
0.422 |
0.007 |
| 54 |
265312_at |
AT2G20240
|
TRM17, TON1 Recruiting Motif 17 |
0.421 |
0.871 |
| 55 |
258935_at |
AT3G10120
|
unknown |
0.421 |
-1.408 |
| 56 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.420 |
-0.848 |
| 57 |
253663_at |
AT4G30160
|
ATVLN4, VLN4, villin 4 |
0.418 |
0.540 |
| 58 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
0.418 |
0.158 |
| 59 |
266439_s_at |
AT2G43200
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.418 |
-0.785 |
| 60 |
257959_at |
AT3G25560
|
AtNIK2, NIK2, NSP-interacting kinase 2 |
0.415 |
-0.247 |
| 61 |
248344_at |
AT5G52280
|
[Myosin heavy chain-related protein] |
0.413 |
0.392 |
| 62 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
0.411 |
0.524 |
| 63 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.411 |
0.950 |
| 64 |
258756_at |
AT3G11960
|
[Cleavage and polyadenylation specificity factor (CPSF) A subunit protein] |
0.407 |
0.819 |
| 65 |
264987_at |
AT1G27030
|
unknown |
0.401 |
0.865 |
| 66 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
0.400 |
-0.511 |
| 67 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.400 |
-0.431 |
| 68 |
245657_at |
AT1G56720
|
[Protein kinase superfamily protein] |
0.400 |
-0.306 |
| 69 |
252482_at |
AT3G46670
|
UGT76E11, UDP-glucosyl transferase 76E11 |
0.397 |
-0.920 |
| 70 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
0.396 |
0.521 |
| 71 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
0.392 |
-0.390 |
| 72 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
0.390 |
0.224 |
| 73 |
253996_at |
AT4G26110
|
ATNAP1;1, ARABIDOPSIS THALIANA NUCLEOSOME ASSEMLY PROTEIN 1;1, NAP1;1, nucleosome assembly protein1;1 |
0.388 |
0.634 |
| 74 |
252268_at |
AT3G49650
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.386 |
0.026 |
| 75 |
251646_at |
AT3G57780
|
unknown |
0.386 |
-0.741 |
| 76 |
256599_at |
AT3G14760
|
unknown |
0.385 |
-0.105 |
| 77 |
252208_at |
AT3G50380
|
[INVOLVED IN: protein localization] |
0.384 |
-0.324 |
| 78 |
245965_at |
AT5G19730
|
[Pectin lyase-like superfamily protein] |
0.383 |
-0.332 |
| 79 |
254235_at |
AT4G23750
|
CRF2, cytokinin response factor 2, TMO3, TARGET OF MONOPTEROS 3 |
0.381 |
0.107 |
| 80 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
0.380 |
0.895 |
| 81 |
248784_at |
AT5G47380
|
unknown |
0.378 |
0.935 |
| 82 |
246231_at |
AT4G37080
|
unknown |
0.377 |
0.708 |
| 83 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.376 |
0.019 |
| 84 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
0.376 |
0.192 |
| 85 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.375 |
-0.515 |
| 86 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
0.370 |
0.014 |
| 87 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.370 |
-0.176 |
| 88 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.366 |
0.585 |
| 89 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.365 |
0.447 |
| 90 |
267089_at |
AT2G38300
|
[myb-like HTH transcriptional regulator family protein] |
0.364 |
-1.530 |
| 91 |
264525_at |
AT1G10060
|
ATBCAT-1, branched-chain amino acid transaminase 1, BCAT-1, branched-chain amino acid transaminase 1, BCAT1, branched-chain amino acid transferase 1 |
0.364 |
0.101 |
| 92 |
255760_at |
AT1G16780
|
AtVHP2;2, VHP2;2 |
0.364 |
0.000 |
| 93 |
245859_at |
AT5G28290
|
ATNEK3, NIMA-related kinase 3, NEK3, NIMA-related kinase 3 |
0.364 |
0.038 |
| 94 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
0.361 |
-0.884 |
| 95 |
246329_at |
AT3G43610
|
[Spc97 / Spc98 family of spindle pole body (SBP) component] |
0.361 |
-0.150 |
| 96 |
265454_at |
AT2G46530
|
ARF11, auxin response factor 11 |
0.360 |
0.111 |
| 97 |
261844_at |
AT1G15940
|
[Tudor/PWWP/MBT superfamily protein] |
0.359 |
-0.045 |
| 98 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.355 |
-0.278 |
| 99 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
0.353 |
0.141 |
| 100 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.349 |
-0.186 |
| 101 |
251714_at |
AT3G56370
|
[Leucine-rich repeat protein kinase family protein] |
0.348 |
0.006 |
| 102 |
266295_at |
AT2G29550
|
TUB7, tubulin beta-7 chain |
0.344 |
0.604 |
| 103 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
0.343 |
-0.729 |
| 104 |
251059_at |
AT5G01810
|
ATPK10, PROTEIN KINASE 10, CIPK15, CBL-interacting protein kinase 15, PKS3, SIP2, SOS3-INTERACTING PROTEIN 2, SNRK3.1, SNF1-RELATED PROTEIN KINASE 3.1 |
0.342 |
-0.210 |
| 105 |
251876_at |
AT3G54280
|
ATBTAF1, BTAF1, CHA16, CHR16, RGD3, ROOT GROWTH DEFECTIVE 3 |
0.342 |
0.101 |
| 106 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
0.341 |
-0.283 |
| 107 |
261471_at |
AT1G14460
|
[AAA-type ATPase family protein] |
0.340 |
-0.193 |
| 108 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.340 |
-0.506 |
| 109 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.338 |
-0.850 |
| 110 |
260638_at |
AT1G62390
|
CLMP1, CLUMPED CHLOROPLASTS 1, Phox2, Phox2 |
0.338 |
1.206 |
| 111 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
0.337 |
1.254 |
| 112 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.334 |
0.338 |
| 113 |
255449_at |
AT4G02820
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.334 |
1.264 |
| 114 |
264782_at |
AT1G08810
|
AtMYB60, myb domain protein 60, MYB60, myb domain protein 60 |
0.332 |
-0.857 |
| 115 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
0.332 |
0.185 |
| 116 |
255891_at |
AT1G17870
|
ATEGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3, EGY3, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 3 |
0.331 |
0.443 |
| 117 |
258293_at |
AT3G23430
|
ATPHO1, ARABIDOPSIS PHOSPHATE 1, PHO1, phosphate 1 |
0.331 |
-0.198 |
| 118 |
261593_at |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.330 |
0.023 |
| 119 |
251852_at |
AT3G54750
|
unknown |
0.330 |
1.218 |
| 120 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.329 |
0.044 |
| 121 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
0.326 |
-0.886 |
| 122 |
259953_at |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
0.325 |
-0.301 |
| 123 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.324 |
-0.412 |
| 124 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
0.324 |
0.006 |
| 125 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
0.323 |
0.635 |
| 126 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
0.323 |
-0.765 |
| 127 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.321 |
-0.452 |
| 128 |
266360_at |
AT2G32250
|
FRS2, FAR1-related sequence 2 |
0.319 |
-0.482 |
| 129 |
252280_at |
AT3G49260
|
iqd21, IQ-domain 21 |
0.317 |
-0.035 |
| 130 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.317 |
0.563 |
| 131 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.316 |
0.050 |
| 132 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.315 |
0.648 |
| 133 |
251734_at |
AT3G56270
|
unknown |
0.313 |
-0.625 |
| 134 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.312 |
0.644 |
| 135 |
261536_at |
AT1G01790
|
ATKEA1, K+ EFFLUX ANTIPORTER 1, KEA1, K+ efflux antiporter 1 |
0.312 |
0.238 |
| 136 |
246738_at |
AT5G27740
|
EMB161, EMBRYO DEFECTIVE 161, EMB251, EMBRYO DEFECTIVE 251, EMB2775, EMBRYO DEFECTIVE 2775, RFC3, replication factor C 3 |
0.311 |
1.092 |
| 137 |
247985_at |
AT5G56790
|
[Protein kinase superfamily protein] |
0.311 |
0.561 |
| 138 |
262491_at |
AT1G21650
|
SECA2 |
0.310 |
-0.423 |
| 139 |
263793_at |
AT2G24630
|
ATCSLC08, ATCSLC8, CELLULOSE-SYNTHASE LIKE C8, CSLC08 |
0.310 |
-0.010 |
| 140 |
263726_at |
AT2G13610
|
ABCG5, ATP-binding cassette G5 |
0.309 |
-0.287 |
| 141 |
245645_at |
AT1G24764
|
ATMAP70-2, microtubule-associated proteins 70-2, MAP70-2, microtubule-associated proteins 70-2 |
0.309 |
0.290 |
| 142 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
0.307 |
-0.246 |
| 143 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
0.305 |
0.746 |
| 144 |
248197_at |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
0.305 |
-0.702 |
| 145 |
253479_at |
AT4G32360
|
[Pyridine nucleotide-disulphide oxidoreductase family protein] |
0.304 |
-0.030 |
| 146 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.303 |
-0.936 |
| 147 |
253974_at |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
0.303 |
-0.627 |
| 148 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.302 |
0.524 |
| 149 |
258675_at |
AT3G08770
|
LTP6, lipid transfer protein 6 |
0.302 |
1.787 |
| 150 |
260347_at |
AT1G69420
|
[DHHC-type zinc finger family protein] |
0.301 |
0.448 |
| 151 |
264932_at |
AT1G61240
|
unknown |
0.300 |
-0.580 |
| 152 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
0.299 |
0.891 |
| 153 |
250751_at |
AT5G05890
|
[UDP-Glycosyltransferase superfamily protein] |
0.298 |
-0.338 |
| 154 |
247977_at |
AT5G56850
|
unknown |
0.297 |
0.507 |
| 155 |
255128_at |
AT4G08310
|
unknown |
0.297 |
1.207 |
| 156 |
262941_at |
AT1G79490
|
EMB2217, embryo defective 2217 |
0.297 |
-0.339 |
| 157 |
255549_at |
AT4G01950
|
ATGPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3, GPAT3, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 3 |
0.297 |
-0.674 |
| 158 |
261796_at |
AT1G30440
|
[Phototropic-responsive NPH3 family protein] |
0.296 |
0.467 |
| 159 |
250955_at |
AT5G03190
|
CPUORF47, conserved peptide upstream open reading frame 47 |
0.295 |
-0.184 |
| 160 |
248283_at |
AT5G52920
|
PKP-BETA1, plastidic pyruvate kinase beta subunit 1, PKP1, PLASTIDIAL PYRUVATE KINASE 1, PKP2, PLASTIDIAL PYRUVATE KINASE 2 |
0.294 |
0.279 |
| 161 |
249821_at |
AT5G23690
|
[Polynucleotide adenylyltransferase family protein] |
0.294 |
0.449 |
| 162 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
0.294 |
0.602 |
| 163 |
263433_at |
AT2G22240
|
ATIPS2, INOSITOL 3-PHOSPHATE SYNTHASE 2, ATMIPS2, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 2, MIPS2, myo-inositol-1-phosphate synthase 2 |
0.293 |
0.109 |
| 164 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.292 |
0.378 |
| 165 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.292 |
-0.603 |
| 166 |
247891_at |
AT5G57960
|
[GTP-binding protein, HflX] |
0.290 |
-0.226 |
| 167 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.287 |
1.057 |
| 168 |
250065_at |
AT5G17910
|
unknown |
0.285 |
0.537 |
| 169 |
262026_at |
AT1G35670
|
ATCDPK2, calcium-dependent protein kinase 2, ATCPK11, CDPK2, calcium-dependent protein kinase 2, CPK11 |
0.285 |
-0.401 |
| 170 |
252023_at |
AT3G52920
|
unknown |
0.284 |
1.067 |
| 171 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.284 |
-0.268 |
| 172 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.284 |
-0.026 |
| 173 |
263127_at |
AT1G78610
|
MSL6, mechanosensitive channel of small conductance-like 6 |
0.282 |
-0.203 |
| 174 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
0.282 |
-0.700 |
| 175 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
0.281 |
0.526 |
| 176 |
253109_at |
AT4G35920
|
MCA1, mid1-complementing activity 1 |
0.280 |
-0.494 |
| 177 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.279 |
-0.842 |
| 178 |
262360_at |
AT1G73080
|
ATPEPR1, PEP1 RECEPTOR 1, PEPR1, PEP1 receptor 1 |
0.278 |
1.026 |
| 179 |
264529_at |
AT1G30820
|
[CTP synthase family protein] |
0.276 |
1.344 |
| 180 |
253407_at |
AT4G32920
|
[glycine-rich protein] |
0.274 |
1.128 |
| 181 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.271 |
-0.111 |
| 182 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
0.270 |
1.459 |
| 183 |
253446_at |
AT4G32620
|
[Enhancer of polycomb-like transcription factor protein] |
0.270 |
0.218 |
| 184 |
259485_at |
AT1G15700
|
ATPC2 |
0.270 |
1.173 |
| 185 |
249702_at |
AT5G35570
|
[O-fucosyltransferase family protein] |
0.270 |
0.372 |
| 186 |
254382_at |
AT4G21890
|
unknown |
0.269 |
-0.631 |
| 187 |
258024_at |
AT3G19360
|
[Zinc finger (CCCH-type) family protein] |
0.269 |
-0.110 |
| 188 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.268 |
0.285 |
| 189 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.267 |
-0.282 |
| 190 |
265453_at |
AT2G46520
|
[cellular apoptosis susceptibility protein, putative / importin-alpha re-exporter, putative] |
0.267 |
1.319 |
| 191 |
258390_at |
AT3G15410
|
[Leucine-rich repeat (LRR) family protein] |
0.267 |
1.067 |
| 192 |
254155_at |
AT4G24480
|
[Protein kinase superfamily protein] |
0.264 |
-0.446 |
| 193 |
252863_at |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
0.263 |
1.196 |
| 194 |
253185_at |
AT4G35240
|
unknown |
0.262 |
-0.085 |
| 195 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.260 |
-0.593 |
| 196 |
259229_at |
AT3G07740
|
ADA2A, homolog of yeast ADA2 2A, ATADA2A, HAC10, HXA02, HXA2 |
0.260 |
-0.135 |
| 197 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.260 |
0.180 |
| 198 |
252954_at |
AT4G38660
|
[Pathogenesis-related thaumatin superfamily protein] |
0.259 |
1.192 |
| 199 |
253495_at |
AT4G31850
|
PGR3, proton gradient regulation 3 |
0.258 |
0.494 |
| 200 |
260308_at |
AT1G70610
|
ABCB26, ATP-binding cassette B26, ATTAP1, transporter associated with antigen processing protein 1, TAP1, transporter associated with antigen processing protein 1 |
0.258 |
1.144 |
| 201 |
251265_at |
AT3G62310
|
[RNA helicase family protein] |
0.257 |
0.140 |
| 202 |
262867_at |
AT1G64960
|
HEB1, hypersensitive to excess boron 1 |
0.257 |
-0.161 |
| 203 |
261314_at |
AT1G52980
|
AtNug2, nuclear/nucleolar GTPase 2, Nug2, nuclear/nucleolar GTPase 2 |
0.256 |
0.786 |
| 204 |
248597_at |
AT5G49160
|
DDM2, DECREASED DNA METHYLATION 2, DMT01, DNA METHYLTRANSFERASE 01, DMT1, DNA METHYLTRANSFERASE 1, MET1, methyltransferase 1, MET2, METHYLTRANSFERASE 2, METI, METHYLTRANSFERASE I |
0.254 |
1.250 |
| 205 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
0.254 |
0.286 |
| 206 |
249093_at |
AT5G43880
|
TRM21, TON1 Recruiting Motif 21 |
0.253 |
0.462 |
| 207 |
248737_at |
AT5G48120
|
MET18, homolog of yeast MET18 |
0.253 |
0.417 |
| 208 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
0.252 |
-0.168 |
| 209 |
266509_at |
AT2G47940
|
DEG2, degradation of periplasmic proteins 2, DEGP2, DEGP protease 2, EMB3117, EMBRYO DEFECTIVE 3117 |
0.251 |
1.299 |
| 210 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.249 |
0.831 |
| 211 |
261768_at |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
0.249 |
0.129 |
| 212 |
249985_at |
AT5G18500
|
[Protein kinase superfamily protein] |
0.248 |
1.105 |
| 213 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.247 |
-0.436 |
| 214 |
253916_at |
AT4G27240
|
[zinc finger (C2H2 type) family protein] |
0.247 |
1.017 |
| 215 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.247 |
-0.481 |
| 216 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.247 |
0.290 |
| 217 |
256650_at |
AT3G13620
|
PUT4, POLYAMINE UPTAKE TRANSPORTER 4 |
0.246 |
-0.071 |
| 218 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-1.411 |
-0.604 |
| 219 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.305 |
-0.399 |
| 220 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
-1.282 |
-0.251 |
| 221 |
260101_at |
AT1G73260
|
ATKTI1, ARABIDOPSIS THALIANA KUNITZ TRYPSIN INHIBITOR 1, KTI1, kunitz trypsin inhibitor 1 |
-1.207 |
0.396 |
| 222 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-1.176 |
-1.027 |
| 223 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.167 |
-0.917 |
| 224 |
267345_at |
AT2G44240
|
unknown |
-1.113 |
-1.136 |
| 225 |
248333_at |
AT5G52390
|
[PAR1 protein] |
-1.077 |
0.719 |
| 226 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
-1.075 |
-0.708 |
| 227 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
-1.061 |
0.375 |
| 228 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-1.050 |
-1.023 |
| 229 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-1.042 |
0.131 |
| 230 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-1.040 |
-1.289 |
| 231 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
-1.018 |
0.484 |
| 232 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.968 |
-0.131 |
| 233 |
258362_at |
AT3G14280
|
unknown |
-0.959 |
-0.824 |
| 234 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.955 |
-0.035 |
| 235 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.947 |
0.370 |
| 236 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
-0.946 |
-0.078 |
| 237 |
256569_at |
AT3G19550
|
unknown |
-0.944 |
-0.780 |
| 238 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
-0.943 |
-0.178 |
| 239 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.928 |
0.036 |
| 240 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.924 |
-0.540 |
| 241 |
261020_at |
AT1G26390
|
[FAD-binding Berberine family protein] |
-0.922 |
-0.140 |
| 242 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.922 |
-0.448 |
| 243 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.903 |
-0.997 |
| 244 |
258209_at |
AT3G14060
|
unknown |
-0.893 |
-0.227 |
| 245 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
-0.892 |
-0.230 |
| 246 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.890 |
-0.281 |
| 247 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
-0.890 |
-0.535 |
| 248 |
266658_at |
AT2G25735
|
unknown |
-0.889 |
-0.540 |
| 249 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.886 |
-0.653 |
| 250 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.883 |
-0.174 |
| 251 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.873 |
-0.727 |
| 252 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.870 |
-0.287 |
| 253 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.864 |
-0.835 |
| 254 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.863 |
-0.475 |
| 255 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.849 |
-0.429 |
| 256 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.846 |
-0.463 |
| 257 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.837 |
0.130 |
| 258 |
247800_at |
AT5G58570
|
unknown |
-0.829 |
0.147 |
| 259 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.829 |
0.452 |
| 260 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-0.829 |
-0.110 |
| 261 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.813 |
-0.532 |
| 262 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-0.810 |
-0.291 |
| 263 |
258203_at |
AT3G13950
|
unknown |
-0.808 |
-0.269 |
| 264 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.804 |
0.326 |
| 265 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
-0.799 |
-0.196 |
| 266 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
-0.799 |
-1.640 |
| 267 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
-0.794 |
-0.452 |
| 268 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
-0.793 |
0.101 |
| 269 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.788 |
-1.172 |
| 270 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.775 |
-1.102 |
| 271 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.772 |
-0.632 |
| 272 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.772 |
-0.587 |
| 273 |
263772_at |
AT2G06255
|
ELF4-L3, ELF4-like 3 |
-0.770 |
0.276 |
| 274 |
256937_at |
AT3G22620
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.768 |
-0.557 |
| 275 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
-0.758 |
-0.455 |
| 276 |
264580_at |
AT1G05340
|
unknown |
-0.753 |
-1.243 |
| 277 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.752 |
-0.587 |
| 278 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-0.751 |
-0.865 |
| 279 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.749 |
-0.184 |
| 280 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.747 |
-1.218 |
| 281 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.742 |
-1.379 |
| 282 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
-0.737 |
-0.746 |
| 283 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.736 |
-0.590 |
| 284 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.735 |
0.033 |
| 285 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.735 |
-0.471 |
| 286 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
-0.734 |
0.414 |
| 287 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.731 |
-0.852 |
| 288 |
247882_at |
AT5G57785
|
unknown |
-0.728 |
0.215 |
| 289 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
-0.727 |
-0.088 |
| 290 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.719 |
-0.475 |
| 291 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.718 |
-0.162 |
| 292 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.715 |
-0.591 |
| 293 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.713 |
-0.723 |
| 294 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.712 |
0.106 |
| 295 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-0.711 |
-0.935 |
| 296 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.710 |
-0.912 |
| 297 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
-0.707 |
-0.321 |
| 298 |
259385_at |
AT1G13470
|
unknown |
-0.696 |
-0.671 |
| 299 |
266017_at |
AT2G18690
|
unknown |
-0.693 |
-0.163 |
| 300 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
-0.692 |
-0.282 |
| 301 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.690 |
-0.891 |
| 302 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.689 |
-0.705 |
| 303 |
265083_at |
AT1G03820
|
unknown |
-0.684 |
0.175 |
| 304 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.682 |
0.185 |
| 305 |
249454_at |
AT5G39520
|
unknown |
-0.678 |
-0.566 |
| 306 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
-0.676 |
0.707 |
| 307 |
264635_at |
AT1G65500
|
unknown |
-0.672 |
0.393 |
| 308 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.668 |
-0.018 |
| 309 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-0.661 |
-0.415 |
| 310 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
-0.661 |
-0.148 |
| 311 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
-0.659 |
-0.648 |
| 312 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.657 |
0.243 |
| 313 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.657 |
0.245 |
| 314 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.654 |
-0.671 |
| 315 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
-0.654 |
-1.017 |
| 316 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.654 |
-0.964 |
| 317 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
-0.653 |
-0.356 |
| 318 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
-0.651 |
-0.310 |
| 319 |
264958_at |
AT1G76960
|
unknown |
-0.650 |
-0.776 |
| 320 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.649 |
-0.698 |
| 321 |
245193_at |
AT1G67810
|
SUFE2, sulfur E2 |
-0.647 |
-0.167 |
| 322 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
-0.645 |
-0.692 |
| 323 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.644 |
-1.042 |
| 324 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.642 |
-0.444 |
| 325 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
-0.642 |
0.195 |
| 326 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.642 |
-0.468 |
| 327 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.639 |
-0.829 |
| 328 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.637 |
-0.388 |
| 329 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.637 |
-0.439 |
| 330 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.637 |
-0.232 |
| 331 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.636 |
-0.138 |
| 332 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.632 |
-1.154 |
| 333 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.630 |
-0.643 |
| 334 |
264590_at |
AT2G17710
|
unknown |
-0.629 |
0.480 |
| 335 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
-0.629 |
0.243 |
| 336 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
-0.628 |
-0.278 |
| 337 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.624 |
0.127 |
| 338 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.624 |
0.990 |
| 339 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-0.622 |
-0.822 |
| 340 |
259489_at |
AT1G15790
|
unknown |
-0.620 |
-0.766 |
| 341 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.620 |
-0.886 |
| 342 |
266352_at |
AT2G01610
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.617 |
-0.287 |
| 343 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.614 |
-1.026 |
| 344 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.613 |
-0.142 |
| 345 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.611 |
-0.522 |
| 346 |
260396_at |
AT1G69720
|
HO3, heme oxygenase 3 |
-0.610 |
-1.214 |
| 347 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.609 |
-0.470 |
| 348 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.608 |
-0.063 |
| 349 |
254853_at |
AT4G12080
|
AHL1, AT-hook motif nuclear-localized protein 1, ATAHL1 |
-0.607 |
-0.005 |
| 350 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.604 |
-0.089 |
| 351 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.604 |
-0.446 |
| 352 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.602 |
-0.723 |
| 353 |
250666_at |
AT5G07100
|
WRKY26, WRKY DNA-binding protein 26 |
-0.601 |
-1.133 |
| 354 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.601 |
-0.303 |
| 355 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.598 |
-0.895 |
| 356 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.596 |
-0.148 |
| 357 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.595 |
0.461 |
| 358 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
-0.591 |
-0.058 |
| 359 |
249377_at |
AT5G40690
|
unknown |
-0.590 |
-0.181 |
| 360 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
-0.589 |
-0.805 |
| 361 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.589 |
-0.950 |
| 362 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.588 |
-0.686 |
| 363 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.588 |
-1.179 |
| 364 |
257697_at |
AT3G12700
|
NANA, NANA |
-0.587 |
0.958 |
| 365 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.587 |
-0.341 |
| 366 |
264005_at |
AT2G22470
|
AGP2, arabinogalactan protein 2, ATAGP2 |
-0.582 |
-0.553 |
| 367 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
-0.581 |
0.219 |
| 368 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.581 |
0.432 |
| 369 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.579 |
-0.610 |
| 370 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.577 |
-0.353 |
| 371 |
257154_at |
AT3G27210
|
unknown |
-0.575 |
-0.778 |
| 372 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.572 |
0.031 |
| 373 |
253827_at |
AT4G28085
|
unknown |
-0.572 |
-0.942 |
| 374 |
267178_at |
AT2G37750
|
unknown |
-0.571 |
-0.416 |
| 375 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.570 |
0.414 |
| 376 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
-0.569 |
-0.270 |
| 377 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.567 |
-0.495 |
| 378 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.567 |
-0.959 |
| 379 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.566 |
1.099 |
| 380 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-0.563 |
-0.399 |
| 381 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-0.560 |
-0.530 |
| 382 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.560 |
-0.577 |
| 383 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.558 |
-0.863 |
| 384 |
258787_at |
AT3G11840
|
PUB24, plant U-box 24 |
-0.556 |
-0.722 |
| 385 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
-0.555 |
-0.808 |
| 386 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
-0.552 |
-0.524 |
| 387 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.551 |
-0.473 |
| 388 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
-0.547 |
-0.805 |
| 389 |
258201_at |
AT3G13910
|
unknown |
-0.546 |
0.721 |
| 390 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.542 |
-0.624 |
| 391 |
259479_at |
AT1G19020
|
unknown |
-0.541 |
-0.031 |
| 392 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
-0.540 |
0.214 |
| 393 |
247071_at |
AT5G66640
|
DAR3, DA1-related protein 3 |
-0.538 |
-0.570 |
| 394 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.534 |
-0.077 |
| 395 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
-0.532 |
-0.569 |
| 396 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.531 |
-0.436 |
| 397 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.526 |
-0.177 |
| 398 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.526 |
-0.343 |
| 399 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.525 |
0.320 |
| 400 |
256256_at |
AT3G11230
|
[Yippee family putative zinc-binding protein] |
-0.525 |
-0.267 |
| 401 |
246200_at |
AT4G37240
|
unknown |
-0.525 |
1.011 |
| 402 |
259661_at |
AT1G55265
|
unknown |
-0.523 |
-0.815 |
| 403 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.522 |
-0.518 |
| 404 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.522 |
-0.168 |
| 405 |
250796_at |
AT5G05300
|
unknown |
-0.520 |
-0.255 |
| 406 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.515 |
0.087 |
| 407 |
248889_at |
AT5G46230
|
unknown |
-0.514 |
-0.222 |
| 408 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.514 |
-1.076 |
| 409 |
265189_at |
AT1G23840
|
unknown |
-0.511 |
-0.378 |
| 410 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.510 |
-1.005 |
| 411 |
246018_at |
AT5G10695
|
unknown |
-0.510 |
-0.520 |
| 412 |
259766_at |
AT1G64360
|
unknown |
-0.506 |
-0.802 |
| 413 |
246197_at |
AT4G37010
|
CEN2, centrin 2 |
-0.503 |
-1.297 |
| 414 |
261700_at |
AT1G32690
|
unknown |
-0.502 |
0.323 |
| 415 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
-0.501 |
-0.194 |
| 416 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.501 |
0.176 |
| 417 |
257074_at |
AT3G19660
|
unknown |
-0.500 |
-0.782 |
| 418 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
-0.500 |
-0.074 |
| 419 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.498 |
-0.407 |
| 420 |
260553_at |
AT2G41800
|
unknown |
-0.498 |
-0.304 |
| 421 |
258059_at |
AT3G29035
|
ANAC059, Arabidopsis NAC domain containing protein 59, ATNAC3, NAC domain containing protein 3, NAC3, NAC domain containing protein 3, ORS1, ORE1 SISTER1 |
-0.497 |
-1.132 |
| 422 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
-0.496 |
-1.111 |
| 423 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
-0.496 |
-0.298 |
| 424 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
-0.491 |
-0.753 |
| 425 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.488 |
-0.064 |
| 426 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.487 |
-0.069 |
| 427 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.487 |
-0.763 |
| 428 |
246001_at |
AT5G20790
|
unknown |
-0.486 |
-0.020 |
| 429 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
-0.484 |
-0.455 |
| 430 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
-0.482 |
0.371 |
| 431 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
-0.481 |
1.317 |
| 432 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
-0.481 |
-0.470 |
| 433 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.479 |
0.120 |