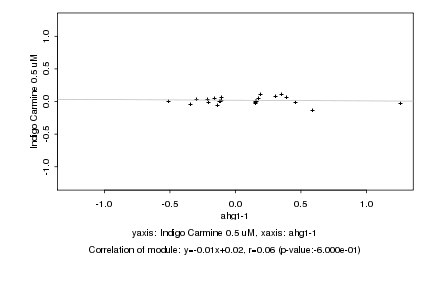
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ahg1-1 |
Log2 signal ratio
Indigo Carmine 0.5 uM |
| 1 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.257 |
-0.016 |
| 2 |
267579_at |
AT2G42000
|
AtMT4a, Arabidopsis thaliana metallothionein 4a |
0.584 |
-0.132 |
| 3 |
259782_at |
AT1G29680
|
unknown |
0.453 |
-0.001 |
| 4 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
0.384 |
0.076 |
| 5 |
258240_at |
AT3G27660
|
OLE3, OLEOSIN 3, OLEO4, oleosin 4 |
0.344 |
0.115 |
| 6 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
0.304 |
0.089 |
| 7 |
249708_at |
AT5G35660
|
[Glycine-rich protein family] |
0.188 |
0.110 |
| 8 |
267210_at |
AT2G30920
|
ATCOQ3, coenzyme Q 3, COQ3, COQ3, coenzyme Q 3, EMB3002, embryo defective 3002 |
0.170 |
0.050 |
| 9 |
259057_at |
AT3G03310
|
ATLCAT3, ARABIDOPSIS LECITHIN:CHOLESTEROL ACYLTRANSFERASE 3, LCAT3, lecithin:cholesterol acyltransferase 3 |
0.158 |
0.006 |
| 10 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.148 |
-0.000 |
| 11 |
254100_at |
AT4G25020
|
[D111/G-patch domain-containing protein] |
0.147 |
0.006 |
| 12 |
256798_at |
AT3G18630
|
ATUNG, uracil dna glycosylase, UNG, uracil dna glycosylase |
0.147 |
-0.024 |
| 13 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.514 |
0.008 |
| 14 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.347 |
-0.042 |
| 15 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.301 |
0.033 |
| 16 |
261471_at |
AT1G14460
|
[AAA-type ATPase family protein] |
-0.215 |
0.042 |
| 17 |
263607_at |
AT2G16270
|
unknown |
-0.206 |
-0.005 |
| 18 |
262462_at |
AT1G50350
|
unknown |
-0.161 |
0.048 |
| 19 |
251652_at |
AT3G57380
|
[Glycosyltransferase family 61 protein] |
-0.144 |
-0.052 |
| 20 |
254845_at |
AT4G11820
|
FKP1, FLAKY POLLEN 1, HMGS, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, MVA1 |
-0.126 |
0.014 |
| 21 |
259466_at |
AT1G19050
|
ARR7, response regulator 7 |
-0.116 |
0.004 |
| 22 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.111 |
0.076 |
| 23 |
254563_at |
AT4G19120
|
ERD3, early-responsive to dehydration 3 |
-0.107 |
0.029 |