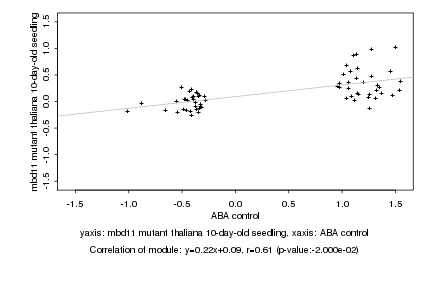
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA control |
Log2 signal ratio
mbd11 mutant thaliana 10-day-old seedling |
| 1 |
255048_at |
AT4G09600
|
GASA3, GAST1 protein homolog 3 |
1.544 |
0.388 |
| 2 |
254440_at |
AT4G21020
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.532 |
0.208 |
| 3 |
254095_at |
AT4G25140
|
OLE1, OLEOSIN 1, OLEO1, oleosin 1 |
1.499 |
1.020 |
| 4 |
246299_at |
AT3G51810
|
AT3, ATEM1, ARABIDOPSIS THALIANA LATE EMBRYOGENESIS ABUNDANT 1, EM1, LATE EMBRYOGENESIS ABUNDANT 1, GEA1, GUANINE NUCLEOTIDE EXCHANGE FACTOR FOR ADP RIBOSYLATION FACTORS 1 |
1.472 |
0.114 |
| 5 |
264079_at |
AT2G28490
|
[RmlC-like cupins superfamily protein] |
1.450 |
0.576 |
| 6 |
266654_at |
AT2G25890
|
[Oleosin family protein] |
1.361 |
0.155 |
| 7 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
1.350 |
0.267 |
| 8 |
250624_at |
AT5G07330
|
unknown |
1.331 |
0.318 |
| 9 |
266393_at |
AT2G41260
|
ATM17, M17 |
1.319 |
0.224 |
| 10 |
256338_at |
AT1G72100
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
1.307 |
0.067 |
| 11 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
1.274 |
0.984 |
| 12 |
263753_at |
AT2G21490
|
LEA, dehydrin LEA |
1.268 |
0.475 |
| 13 |
263385_at |
AT2G40170
|
ATEM6, ARABIDOPSIS EARLY METHIONINE-LABELLED 6, EM6, EARLY METHIONINE-LABELLED 6, GEA6, LATE EMBRYOGENESIS ABUNDANT 6 |
1.252 |
-0.115 |
| 14 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
1.251 |
0.139 |
| 15 |
264494_at |
AT1G27461
|
unknown |
1.244 |
0.080 |
| 16 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
1.192 |
0.364 |
| 17 |
252019_at |
AT3G53040
|
[late embryogenesis abundant protein, putative / LEA protein, putative] |
1.152 |
0.141 |
| 18 |
256354_at |
AT1G54870
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.144 |
0.155 |
| 19 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
1.141 |
0.636 |
| 20 |
250882_at |
AT5G04000
|
unknown |
1.134 |
0.435 |
| 21 |
253767_at |
AT4G28520
|
CRC, CRUCIFERIN C, CRU3, cruciferin 3 |
1.129 |
0.894 |
| 22 |
252137_at |
AT3G50980
|
XERO1, dehydrin xero 1 |
1.113 |
0.020 |
| 23 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
1.105 |
0.880 |
| 24 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
1.079 |
0.105 |
| 25 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
1.078 |
0.582 |
| 26 |
260088_at |
AT1G73190
|
ALPHA-TIP, ALPHA-TONOPLAST INTRINSIC PROTEIN, TIP3;1 |
1.057 |
0.373 |
| 27 |
256827_at |
AT3G18570
|
[Oleosin family protein] |
1.055 |
0.256 |
| 28 |
259167_at |
AT3G01570
|
[Oleosin family protein] |
1.041 |
0.685 |
| 29 |
260458_at |
AT1G68250
|
unknown |
1.041 |
0.064 |
| 30 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
1.009 |
0.512 |
| 31 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.974 |
0.348 |
| 32 |
266143_at |
AT2G38905
|
[Low temperature and salt responsive protein family] |
0.967 |
0.280 |
| 33 |
259217_at |
AT3G03620
|
[MATE efflux family protein] |
0.956 |
0.295 |
| 34 |
265050_at |
AT1G52070
|
[Mannose-binding lectin superfamily protein] |
-1.016 |
-0.184 |
| 35 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
-0.881 |
-0.033 |
| 36 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.665 |
-0.164 |
| 37 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.554 |
0.005 |
| 38 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.545 |
-0.195 |
| 39 |
247878_at |
AT5G57760
|
unknown |
-0.515 |
0.270 |
| 40 |
254798_at |
AT4G13050
|
[Acyl-ACP thioesterase] |
-0.493 |
-0.145 |
| 41 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.479 |
0.049 |
| 42 |
265048_at |
AT1G52050
|
[Mannose-binding lectin superfamily protein] |
-0.471 |
0.043 |
| 43 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.468 |
-0.159 |
| 44 |
253823_at |
AT4G28030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.455 |
0.032 |
| 45 |
247943_at |
AT5G57170
|
[Tautomerase/MIF superfamily protein] |
-0.440 |
0.205 |
| 46 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
-0.430 |
-0.176 |
| 47 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.420 |
-0.261 |
| 48 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.417 |
0.243 |
| 49 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
-0.408 |
0.080 |
| 50 |
259752_at |
AT1G71040
|
LPR2, Low Phosphate Root2 |
-0.402 |
0.053 |
| 51 |
252876_at |
AT4G39970
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.401 |
0.110 |
| 52 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.382 |
-0.092 |
| 53 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
-0.376 |
-0.002 |
| 54 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
-0.373 |
-0.135 |
| 55 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.372 |
0.183 |
| 56 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.362 |
0.157 |
| 57 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
-0.352 |
-0.206 |
| 58 |
250262_at |
AT5G13410
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.350 |
0.110 |
| 59 |
258662_at |
AT3G02900
|
unknown |
-0.346 |
0.132 |
| 60 |
245321_at |
AT4G15545
|
unknown |
-0.339 |
-0.119 |
| 61 |
247544_at |
AT5G61670
|
unknown |
-0.334 |
-0.049 |
| 62 |
246997_at |
AT5G67390
|
unknown |
-0.331 |
-0.087 |
| 63 |
248624_at |
AT5G48790
|
unknown |
-0.323 |
-0.095 |
| 64 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.297 |
0.113 |
| 65 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.285 |
0.029 |