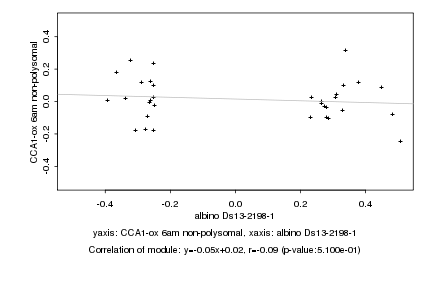
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
albino Ds13-2198-1 |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
256631_at |
AT3G28320
|
unknown |
0.506 |
-0.242 |
| 2 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.480 |
-0.078 |
| 3 |
256381_at |
AT1G66850
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.447 |
0.089 |
| 4 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.376 |
0.123 |
| 5 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
0.338 |
0.315 |
| 6 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.330 |
0.102 |
| 7 |
252322_at |
AT3G48550
|
unknown |
0.328 |
-0.053 |
| 8 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
0.310 |
0.049 |
| 9 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.305 |
0.025 |
| 10 |
250869_at |
AT5G03840
|
TFL-1, TERMINAL FLOWER 1, TFL1, TERMINAL FLOWER 1 |
0.284 |
-0.099 |
| 11 |
251665_at |
AT3G57040
|
ARR9, response regulator 9, ATRR4, RESPONSE REGULATOR 4 |
0.279 |
-0.098 |
| 12 |
261224_at |
AT1G20160
|
ATSBT5.2 |
0.278 |
-0.031 |
| 13 |
254145_at |
AT4G24700
|
unknown |
0.272 |
-0.025 |
| 14 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.263 |
0.002 |
| 15 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.262 |
-0.009 |
| 16 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
0.232 |
0.030 |
| 17 |
259001_at |
AT3G01960
|
unknown |
0.228 |
-0.093 |
| 18 |
253582_at |
AT4G30670
|
[Putative membrane lipoprotein] |
-0.395 |
0.007 |
| 19 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
-0.366 |
0.183 |
| 20 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-0.340 |
0.024 |
| 21 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.325 |
0.254 |
| 22 |
267548_at |
AT2G32660
|
AtRLP22, receptor like protein 22, RLP22, receptor like protein 22 |
-0.308 |
-0.178 |
| 23 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
-0.290 |
0.123 |
| 24 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.277 |
-0.167 |
| 25 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
-0.271 |
-0.090 |
| 26 |
256781_at |
AT3G13650
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.265 |
-0.001 |
| 27 |
248605_at |
AT5G49410
|
unknown |
-0.264 |
0.124 |
| 28 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
-0.264 |
0.008 |
| 29 |
266017_at |
AT2G18690
|
unknown |
-0.254 |
-0.176 |
| 30 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-0.254 |
0.028 |
| 31 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.254 |
0.235 |
| 32 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.252 |
0.101 |
| 33 |
265796_at |
AT2G35730
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.251 |
-0.024 |