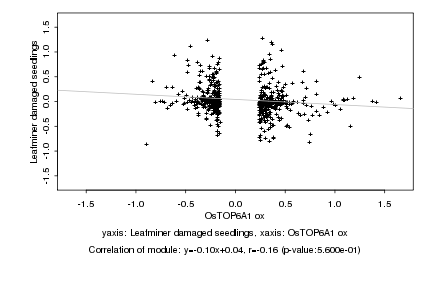
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
OsTOP6A1 ox |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
255257_at |
AT4G05050
|
UBQ11, ubiquitin 11 |
1.655 |
0.023 |
| 2 |
265611_at |
AT2G25510
|
unknown |
1.416 |
-0.009 |
| 3 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
1.368 |
0.001 |
| 4 |
245448_at |
AT4G16860
|
RPP4, recognition of peronospora parasitica 4 |
1.242 |
0.490 |
| 5 |
248944_at |
AT5G45500
|
[RNI-like superfamily protein] |
1.181 |
0.077 |
| 6 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
1.150 |
-0.503 |
| 7 |
265831_at |
AT2G14460
|
unknown |
1.120 |
0.043 |
| 8 |
263023_at |
AT1G23960
|
unknown |
1.088 |
0.040 |
| 9 |
253894_at |
AT4G27150
|
AT2S2, SESA2, seed storage albumin 2 |
1.076 |
0.021 |
| 10 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
1.076 |
0.048 |
| 11 |
256870_at |
AT3G26300
|
CYP71B34, cytochrome P450, family 71, subfamily B, polypeptide 34 |
1.051 |
-0.142 |
| 12 |
252659_at |
AT3G44430
|
unknown |
0.997 |
-0.076 |
| 13 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.976 |
-0.052 |
| 14 |
262908_at |
AT1G59900
|
AT-E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit, E1 ALPHA, pyruvate dehydrogenase complex E1 alpha subunit |
0.958 |
0.004 |
| 15 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
0.916 |
-0.218 |
| 16 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.876 |
-0.116 |
| 17 |
261942_at |
AT1G22590
|
AGL87, AGAMOUS-like 87 |
0.843 |
-0.277 |
| 18 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.814 |
0.159 |
| 19 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
0.809 |
-0.187 |
| 20 |
256021_at |
AT1G58270
|
ZW9 |
0.807 |
0.411 |
| 21 |
254931_at |
AT4G11460
|
CRK30, cysteine-rich RLK (RECEPTOR-like protein kinase) 30 |
0.766 |
-0.270 |
| 22 |
264166_at |
AT1G65370
|
[TRAF-like family protein] |
0.757 |
-0.099 |
| 23 |
246001_at |
AT5G20790
|
unknown |
0.748 |
-0.646 |
| 24 |
257057_at |
AT3G15310
|
unknown |
0.743 |
-0.810 |
| 25 |
259573_at |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
0.728 |
-0.383 |
| 26 |
246888_at |
AT5G26270
|
unknown |
0.708 |
0.273 |
| 27 |
245460_at |
AT4G16990
|
RLM3, RESISTANCE TO LEPTOSPHAERIA MACULANS 3 |
0.705 |
-0.069 |
| 28 |
255160_at |
AT4G07820
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.704 |
0.098 |
| 29 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.693 |
-0.251 |
| 30 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.684 |
0.622 |
| 31 |
248079_at |
AT5G55790
|
unknown |
0.677 |
-0.024 |
| 32 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
0.674 |
0.034 |
| 33 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
0.665 |
0.390 |
| 34 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.652 |
-0.275 |
| 35 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.625 |
-0.233 |
| 36 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
0.622 |
-0.003 |
| 37 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
0.574 |
0.005 |
| 38 |
262010_at |
AT1G35612
|
[expressed protein] |
0.572 |
-0.027 |
| 39 |
260549_at |
AT2G43535
|
[Scorpion toxin-like knottin superfamily protein] |
0.572 |
0.382 |
| 40 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.555 |
-0.043 |
| 41 |
263873_at |
AT2G21860
|
[violaxanthin de-epoxidase-related] |
0.548 |
-0.181 |
| 42 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.540 |
-0.505 |
| 43 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.539 |
-0.040 |
| 44 |
251347_at |
AT3G61010
|
[Ferritin/ribonucleotide reductase-like family protein] |
0.538 |
-0.017 |
| 45 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
0.521 |
-0.016 |
| 46 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.516 |
-0.121 |
| 47 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.514 |
-0.475 |
| 48 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.512 |
-0.005 |
| 49 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
0.512 |
-0.093 |
| 50 |
254724_at |
AT4G13630
|
unknown |
0.512 |
-0.046 |
| 51 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.508 |
-0.496 |
| 52 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.505 |
0.136 |
| 53 |
248183_at |
AT5G54040
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.503 |
-0.009 |
| 54 |
249010_at |
AT5G44580
|
unknown |
0.502 |
-0.018 |
| 55 |
264958_at |
AT1G76960
|
unknown |
0.500 |
0.352 |
| 56 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
0.483 |
0.103 |
| 57 |
245411_at |
AT4G17240
|
unknown |
0.477 |
0.299 |
| 58 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
0.475 |
0.026 |
| 59 |
266766_at |
AT2G46880
|
ATPAP14, PAP14, purple acid phosphatase 14 |
0.473 |
-0.028 |
| 60 |
248954_at |
AT5G45420
|
maMYB, membrane anchored MYB |
0.473 |
0.035 |
| 61 |
254683_at |
AT4G13800
|
unknown |
0.471 |
0.221 |
| 62 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.470 |
0.120 |
| 63 |
245456_at |
AT4G16950
|
RPP5, RECOGNITION OF PERONOSPORA PARASITICA 5 |
0.469 |
0.133 |
| 64 |
246260_at |
AT1G31820
|
PUT1, POLYAMINE UPTAKE TRANSPORTER 1 |
0.467 |
-0.005 |
| 65 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
0.466 |
-0.028 |
| 66 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.466 |
0.715 |
| 67 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.462 |
0.238 |
| 68 |
250755_at |
AT5G05750
|
[DNAJ heat shock N-terminal domain-containing protein] |
0.461 |
0.065 |
| 69 |
266384_at |
AT2G14660
|
unknown |
0.461 |
-0.342 |
| 70 |
257423_at |
AT1G62010
|
[Mitochondrial transcription termination factor family protein] |
0.455 |
-0.001 |
| 71 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.454 |
1.034 |
| 72 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
0.453 |
0.212 |
| 73 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.452 |
0.029 |
| 74 |
262873_at |
AT1G64700
|
unknown |
0.447 |
-0.041 |
| 75 |
255044_at |
AT4G09680
|
ATCTC1, CTC1, conserved telomere maintenance component 1 |
0.447 |
-0.079 |
| 76 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.445 |
-0.038 |
| 77 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
0.441 |
-0.381 |
| 78 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.440 |
-0.160 |
| 79 |
245079_at |
AT2G23330
|
[copia-like retrotransposon family, has a 3.9e-195 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
0.435 |
0.124 |
| 80 |
247755_at |
AT5G59090
|
ATSBT4.12, subtilase 4.12, SBT4.12, subtilase 4.12 |
0.430 |
0.064 |
| 81 |
245799_at |
AT1G45616
|
AtRLP6, receptor like protein 6, RLP6, receptor like protein 6 |
0.429 |
-0.011 |
| 82 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
0.427 |
0.004 |
| 83 |
260012_at |
AT1G67865
|
unknown |
0.427 |
0.169 |
| 84 |
259399_at |
AT1G17710
|
AtPEPC1, Arabidopsis thaliana phosphoethanolamine/phosphocholine phosphatase 1, PEPC1, phosphoethanolamine/phosphocholine phosphatase 1 |
0.426 |
-0.316 |
| 85 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.424 |
0.384 |
| 86 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
0.422 |
-0.008 |
| 87 |
254455_at |
AT4G21140
|
unknown |
0.421 |
-0.010 |
| 88 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
0.418 |
-0.038 |
| 89 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.416 |
0.087 |
| 90 |
265302_at |
AT2G13970
|
[Mutator-like transposase family, has a 4.1e-39 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
0.413 |
-0.055 |
| 91 |
252444_at |
AT3G46980
|
PHT4;3, phosphate transporter 4;3 |
0.413 |
-0.068 |
| 92 |
266235_at |
AT2G02360
|
AtPP2-B10, phloem protein 2-B10, PP2-B10, phloem protein 2-B10 |
0.413 |
0.231 |
| 93 |
258045_at |
AT3G21280
|
UBP7, ubiquitin-specific protease 7 |
0.409 |
-0.022 |
| 94 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
0.407 |
-0.010 |
| 95 |
265439_at |
AT2G21045
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.406 |
-0.028 |
| 96 |
250931_at |
AT5G03200
|
LUL1, LOG2-LIKE UBIQUITIN LIGASE1 |
0.406 |
0.322 |
| 97 |
263597_at |
AT2G01870
|
unknown |
0.404 |
-0.242 |
| 98 |
245106_at |
AT2G41650
|
unknown |
0.402 |
-0.026 |
| 99 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.401 |
-0.111 |
| 100 |
251493_at |
AT3G59300
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.397 |
-0.141 |
| 101 |
264454_at |
AT1G10320
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.397 |
-0.016 |
| 102 |
254437_s_at |
AT3G06433
|
[pseudogene] |
0.397 |
-0.097 |
| 103 |
262028_at |
AT1G35560
|
AtTCP23, TCP23, TCP domain protein 23 |
0.396 |
-0.132 |
| 104 |
251293_at |
AT3G61930
|
unknown |
0.396 |
0.635 |
| 105 |
249258_at |
AT5G41650
|
[Lactoylglutathione lyase / glyoxalase I family protein] |
0.395 |
0.039 |
| 106 |
251165_at |
AT3G63340
|
[Protein phosphatase 2C family protein] |
0.391 |
-0.139 |
| 107 |
256920_at |
AT3G18980
|
ETP1, EIN2 targeting protein1 |
0.390 |
-0.085 |
| 108 |
251600_at |
AT3G57710
|
[Protein kinase superfamily protein] |
0.384 |
-0.019 |
| 109 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.381 |
-0.720 |
| 110 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
0.377 |
-0.035 |
| 111 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.376 |
-0.464 |
| 112 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.375 |
0.198 |
| 113 |
248759_at |
AT5G47610
|
[RING/U-box superfamily protein] |
0.374 |
-0.733 |
| 114 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.372 |
-0.343 |
| 115 |
264874_at |
AT1G24240
|
[Ribosomal protein L19 family protein] |
0.370 |
0.078 |
| 116 |
254770_at |
AT4G13340
|
LRX3, leucine-rich repeat/extensin 3 |
0.369 |
-0.104 |
| 117 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.369 |
0.472 |
| 118 |
251967_at |
AT3G53390
|
[Transducin/WD40 repeat-like superfamily protein] |
0.368 |
-0.009 |
| 119 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.367 |
-0.480 |
| 120 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.365 |
1.152 |
| 121 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
0.364 |
0.023 |
| 122 |
248212_at |
AT5G54020
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.362 |
0.019 |
| 123 |
248940_at |
AT5G45400
|
ATRPA70C, RPA70C |
0.360 |
-0.108 |
| 124 |
261858_at |
AT1G50570
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.359 |
0.111 |
| 125 |
252419_at |
AT3G47510
|
unknown |
0.358 |
0.198 |
| 126 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.358 |
0.141 |
| 127 |
264137_at |
AT1G78960
|
ATLUP2, lupeol synthase 2, LUP2, lupeol synthase 2 |
0.357 |
-0.058 |
| 128 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
0.356 |
0.053 |
| 129 |
267264_at |
AT2G22970
|
SCPL11, serine carboxypeptidase-like 11 |
0.356 |
0.011 |
| 130 |
247360_at |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
0.355 |
1.204 |
| 131 |
261402_at |
AT1G79670
|
RFO1, RESISTANCE TO FUSARIUM OXYSPORUM 1, WAKL22 |
0.352 |
-0.007 |
| 132 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
0.351 |
-0.086 |
| 133 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
0.350 |
-0.313 |
| 134 |
245412_at |
AT4G17280
|
[Auxin-responsive family protein] |
0.349 |
0.027 |
| 135 |
257334_at |
ATMG01370
|
ORF111D |
0.347 |
0.079 |
| 136 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.346 |
0.603 |
| 137 |
267497_at |
AT2G30540
|
[Thioredoxin superfamily protein] |
0.346 |
-0.049 |
| 138 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.345 |
-0.522 |
| 139 |
257554_at |
AT3G24890
|
ATVAMP728, VAMP728, vesicle-associated membrane protein 728 |
0.345 |
0.002 |
| 140 |
245055_at |
AT2G26470
|
unknown |
0.344 |
-0.089 |
| 141 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
0.344 |
-0.203 |
| 142 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
0.344 |
-0.425 |
| 143 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.343 |
0.866 |
| 144 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.342 |
-0.796 |
| 145 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.341 |
0.067 |
| 146 |
255939_at |
AT1G12730
|
[GPI transamidase subunit PIG-U] |
0.341 |
-0.088 |
| 147 |
254600_at |
AT4G19010
|
[AMP-dependent synthetase and ligase family protein] |
0.340 |
0.079 |
| 148 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
0.340 |
0.967 |
| 149 |
245479_at |
AT4G16140
|
[proline-rich family protein] |
0.338 |
-0.205 |
| 150 |
252160_at |
AT3G50570
|
[hydroxyproline-rich glycoprotein family protein] |
0.338 |
-0.258 |
| 151 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
0.337 |
-0.068 |
| 152 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.336 |
0.369 |
| 153 |
252654_at |
AT3G44740
|
[Class II aaRS and biotin synthetases superfamily protein] |
0.336 |
-0.027 |
| 154 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.335 |
0.267 |
| 155 |
261123_at |
AT1G62860
|
|
0.330 |
-0.021 |
| 156 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.329 |
-0.036 |
| 157 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
0.327 |
0.576 |
| 158 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
0.326 |
-0.106 |
| 159 |
255597_at |
AT4G01730
|
[DHHC-type zinc finger family protein] |
0.325 |
-0.044 |
| 160 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
0.325 |
-0.145 |
| 161 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.324 |
-0.553 |
| 162 |
257421_at |
AT1G12030
|
unknown |
0.322 |
-0.375 |
| 163 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.318 |
-0.081 |
| 164 |
247729_at |
AT5G59530
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.318 |
0.291 |
| 165 |
251727_at |
AT3G56290
|
unknown |
0.315 |
-0.250 |
| 166 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.314 |
-0.287 |
| 167 |
246699_at |
AT5G27990
|
[Pre-rRNA-processing protein TSR2, conserved region] |
0.314 |
0.022 |
| 168 |
246079_at |
AT5G20450
|
unknown |
0.313 |
-0.268 |
| 169 |
258137_at |
AT3G24515
|
UBC37, ubiquitin-conjugating enzyme 37 |
0.313 |
0.095 |
| 170 |
253239_at |
AT4G34500
|
[Protein kinase superfamily protein] |
0.313 |
-0.082 |
| 171 |
257322_at |
ATMG01180
|
ORF111B |
0.310 |
0.051 |
| 172 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
0.310 |
-0.059 |
| 173 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.309 |
0.286 |
| 174 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
0.308 |
0.668 |
| 175 |
258856_at |
AT3G02040
|
AtGDPD1, GDPD1, Glycerophosphodiester phosphodiesterase 1, SRG3, senescence-related gene 3 |
0.307 |
-0.234 |
| 176 |
246818_at |
AT5G27270
|
EMB976, EMBRYO DEFECTIVE 976 |
0.307 |
-0.306 |
| 177 |
249592_at |
AT5G37890
|
[Protein with RING/U-box and TRAF-like domains] |
0.306 |
-0.099 |
| 178 |
251231_at |
AT3G62760
|
ATGSTF13 |
0.303 |
-0.019 |
| 179 |
264587_at |
AT1G05200
|
ATGLR3.4, glutamate receptor 3.4, GLR3.4, glutamate receptor 3.4, GLUR3 |
0.303 |
-0.180 |
| 180 |
263515_at |
AT2G21640
|
unknown |
0.302 |
0.136 |
| 181 |
248125_at |
AT5G54740
|
SESA5, seed storage albumin 5 |
0.302 |
-0.053 |
| 182 |
267406_at |
AT2G34780
|
EMB1611, EMBRYO DEFECTIVE 1611, MEE22, MATERNAL EFFECT EMBRYO ARREST 22 |
0.301 |
-0.172 |
| 183 |
258107_at |
AT3G23560
|
ALF5, ABERRANT LATERAL ROOT FORMATION 5 |
0.300 |
0.132 |
| 184 |
263609_at |
AT2G16250
|
[Leucine-rich repeat protein kinase family protein] |
0.299 |
-0.227 |
| 185 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.298 |
-0.404 |
| 186 |
250696_at |
AT5G06790
|
unknown |
0.298 |
-0.735 |
| 187 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
0.298 |
0.274 |
| 188 |
259686_at |
AT1G63100
|
[GRAS family transcription factor] |
0.298 |
-0.010 |
| 189 |
250771_at |
AT5G05400
|
[LRR and NB-ARC domains-containing disease resistance protein] |
0.297 |
0.016 |
| 190 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.295 |
-0.281 |
| 191 |
257410_at |
AT1G24340
|
EMB2421, EMBRYO DEFECTIVE 2421, EMB260, EMBRYO DEFECTIVE 260 |
0.293 |
-0.128 |
| 192 |
248736_at |
AT5G48110
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.292 |
0.039 |
| 193 |
266064_at |
AT2G18780
|
[F-box and associated interaction domains-containing protein] |
0.292 |
0.036 |
| 194 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.292 |
0.558 |
| 195 |
260517_at |
AT1G51420
|
ATSPP1, SUCROSE-PHOSPHATASE 1, SPP1, sucrose-phosphatase 1 |
0.291 |
0.003 |
| 196 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.291 |
-0.373 |
| 197 |
251143_at |
AT5G01220
|
SQD2, sulfoquinovosyldiacylglycerol 2 |
0.290 |
-0.316 |
| 198 |
252829_at |
AT4G40060
|
ATHB-16, ATHB16, homeobox protein 16, HB16, homeobox protein 16 |
0.290 |
-0.060 |
| 199 |
257978_at |
AT3G20860
|
ATNEK5, NIMA-related kinase 5, NEK5, NIMA-related kinase 5 |
0.290 |
0.310 |
| 200 |
245297_at |
AT4G16510
|
[YbaK/aminoacyl-tRNA synthetase-associated domain] |
0.290 |
-0.032 |
| 201 |
256014_at |
AT1G19200
|
unknown |
0.290 |
0.126 |
| 202 |
251728_at |
AT3G56300
|
[Cysteinyl-tRNA synthetase, class Ia family protein] |
0.289 |
-0.151 |
| 203 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.289 |
0.807 |
| 204 |
249639_at |
AT5G36930
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.289 |
-0.170 |
| 205 |
256340_at |
AT1G72070
|
[Chaperone DnaJ-domain superfamily protein] |
0.287 |
0.149 |
| 206 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.285 |
-0.003 |
| 207 |
262561_at |
AT1G34340
|
[alpha/beta-Hydrolases superfamily protein] |
0.285 |
0.144 |
| 208 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.284 |
0.010 |
| 209 |
259379_at |
AT3G16350
|
[Homeodomain-like superfamily protein] |
0.284 |
0.156 |
| 210 |
264501_at |
AT1G09390
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.284 |
-0.173 |
| 211 |
247055_at |
AT5G66740
|
unknown |
0.283 |
-0.594 |
| 212 |
252537_at |
AT3G45710
|
[Major facilitator superfamily protein] |
0.281 |
-0.018 |
| 213 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.280 |
-0.071 |
| 214 |
264183_at |
AT1G65380
|
AtRLP10, Receptor Like Protein 10, CLV2, clavata 2 |
0.280 |
-0.269 |
| 215 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.280 |
0.671 |
| 216 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.279 |
0.843 |
| 217 |
266729_at |
AT2G03130
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
0.279 |
-0.011 |
| 218 |
253156_at |
AT4G35730
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.279 |
-0.050 |
| 219 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.278 |
0.814 |
| 220 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
0.276 |
-0.018 |
| 221 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.275 |
-0.030 |
| 222 |
245453_at |
AT4G16900
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.274 |
0.010 |
| 223 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
0.274 |
0.141 |
| 224 |
248192_at |
AT5G54140
|
ILL3, IAA-leucine-resistant (ILR1)-like 3 |
0.273 |
0.196 |
| 225 |
246633_at |
AT1G29720
|
[Leucine-rich repeat transmembrane protein kinase] |
0.272 |
-0.236 |
| 226 |
254587_at |
AT4G19520
|
[disease resistance protein (TIR-NBS-LRR class) family] |
0.269 |
0.036 |
| 227 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.268 |
0.661 |
| 228 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
0.268 |
1.277 |
| 229 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
0.266 |
-0.107 |
| 230 |
262366_at |
AT1G72890
|
[Disease resistance protein (TIR-NBS class)] |
0.266 |
-0.189 |
| 231 |
252730_at |
AT3G43110
|
unknown |
0.266 |
-0.230 |
| 232 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
0.265 |
0.654 |
| 233 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.264 |
-0.049 |
| 234 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.264 |
0.783 |
| 235 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.263 |
0.287 |
| 236 |
256361_at |
AT1G66520
|
pde194, pigment defective 194 |
0.263 |
-0.170 |
| 237 |
262897_at |
AT1G59840
|
CCB4, cofactor assembly of complex C |
0.263 |
-0.196 |
| 238 |
248181_at |
AT5G54290
|
CcdA |
0.263 |
-0.090 |
| 239 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.262 |
-0.045 |
| 240 |
246808_at |
AT5G27110
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.262 |
-0.087 |
| 241 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.262 |
0.094 |
| 242 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.261 |
0.183 |
| 243 |
251966_at |
AT3G53380
|
LecRK-VIII.1, L-type lectin receptor kinase VIII.1 |
0.261 |
-0.220 |
| 244 |
254909_at |
AT4G11210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.260 |
-0.054 |
| 245 |
254928_at |
AT4G11410
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.260 |
-0.154 |
| 246 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.260 |
-0.351 |
| 247 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.259 |
-0.335 |
| 248 |
257013_at |
AT3G26922
|
[F-box/RNI-like superfamily protein] |
0.259 |
-0.048 |
| 249 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.259 |
-0.526 |
| 250 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.258 |
-0.118 |
| 251 |
250740_at |
AT5G05760
|
ATSED5, ATSYP31, SED5, T-SNARE SED 5, SYP31, syntaxin of plants 31 |
0.258 |
0.107 |
| 252 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.256 |
0.749 |
| 253 |
252637_at |
AT3G44530
|
HIRA, homolog of histone chaperone HIRA |
0.256 |
-0.057 |
| 254 |
246393_at |
AT1G58150
|
unknown |
0.255 |
-0.200 |
| 255 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
0.255 |
0.228 |
| 256 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.254 |
0.560 |
| 257 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
0.254 |
0.035 |
| 258 |
254638_at |
AT4G18740
|
[Rho termination factor] |
0.254 |
-0.339 |
| 259 |
254476_at |
AT4G20410
|
GAMMA-SNAP, GAMMA-SOLUBLE NSF ATTACHMENT PROTEIN, GSNAP, gamma-soluble NSF attachment protein |
0.253 |
0.109 |
| 260 |
257112_at |
AT3G20120
|
CYP705A21, cytochrome P450, family 705, subfamily A, polypeptide 21 |
0.252 |
-0.076 |
| 261 |
257432_at |
AT2G21850
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.251 |
0.061 |
| 262 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.251 |
-0.032 |
| 263 |
260966_at |
AT1G12220
|
RPS5, RESISTANT TO P. SYRINGAE 5 |
0.250 |
-0.040 |
| 264 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.249 |
-0.005 |
| 265 |
259956_at |
AT1G75110
|
RRA2, REDUCED RESIDUAL ARABINOSE 2 |
0.249 |
-0.082 |
| 266 |
248762_at |
AT5G47455
|
unknown |
0.248 |
0.098 |
| 267 |
253530_at |
AT4G31530
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.248 |
-0.057 |
| 268 |
246803_at |
AT5G26980
|
ATSYP41, ATTLG2A, SYP41, syntaxin of plants 41 |
0.248 |
0.067 |
| 269 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.247 |
-0.387 |
| 270 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
0.247 |
-0.678 |
| 271 |
245386_at |
AT4G14010
|
RALFL32, ralf-like 32 |
0.247 |
-0.018 |
| 272 |
252163_at |
AT3G50610
|
unknown |
0.247 |
0.007 |
| 273 |
258501_at |
AT3G06780
|
[glycine-rich protein] |
0.247 |
0.076 |
| 274 |
250453_at |
AT5G10620
|
[methyltransferases] |
0.247 |
-0.101 |
| 275 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
0.246 |
-0.780 |
| 276 |
248942_at |
AT5G45480
|
unknown |
0.246 |
0.139 |
| 277 |
253856_at |
AT4G28100
|
unknown |
0.245 |
-0.217 |
| 278 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.245 |
0.009 |
| 279 |
246286_at |
AT1G31910
|
[GHMP kinase family protein] |
0.245 |
-0.042 |
| 280 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.244 |
-0.415 |
| 281 |
265011_at |
AT1G24490
|
ALB4, ALBINA 4, ARTEMIS, ARABIDOPSIS THALIANA ENVELOPE MEMBRANE INTEGRASE |
0.243 |
-0.117 |
| 282 |
250530_at |
AT5G08630
|
[DDT domain-containing protein] |
0.243 |
0.008 |
| 283 |
258638_at |
AT3G07950
|
[rhomboid protein-related] |
0.242 |
0.013 |
| 284 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.241 |
0.675 |
| 285 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.240 |
-0.279 |
| 286 |
252490_at |
AT3G46720
|
[UDP-Glycosyltransferase superfamily protein] |
0.240 |
-0.019 |
| 287 |
244936_at |
ATCG01100
|
NDHA |
0.239 |
-0.072 |
| 288 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.239 |
-0.414 |
| 289 |
246071_at |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
0.239 |
-0.175 |
| 290 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.236 |
0.730 |
| 291 |
262884_at |
AT1G64720
|
CP5 |
0.236 |
-0.043 |
| 292 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.236 |
0.486 |
| 293 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
0.236 |
-0.713 |
| 294 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.235 |
-0.358 |
| 295 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
0.235 |
0.408 |
| 296 |
245977_at |
AT5G13110
|
G6PD2, glucose-6-phosphate dehydrogenase 2 |
0.234 |
-0.112 |
| 297 |
248951_at |
AT5G45550
|
MOB1-like, MOB1-like |
0.234 |
-0.114 |
| 298 |
249535_at |
AT5G38820
|
[Transmembrane amino acid transporter family protein] |
0.234 |
-0.042 |
| 299 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.234 |
-0.261 |
| 300 |
254388_at |
AT4G21860
|
MSRB2, methionine sulfoxide reductase B 2 |
0.233 |
-0.035 |
| 301 |
254574_at |
AT4G19430
|
unknown |
-0.894 |
-0.848 |
| 302 |
252345_at |
AT3G48640
|
unknown |
-0.834 |
0.420 |
| 303 |
250135_at |
AT5G15360
|
unknown |
-0.810 |
-0.011 |
| 304 |
245703_at |
AT5G04380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.759 |
0.002 |
| 305 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.736 |
0.001 |
| 306 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.715 |
-0.013 |
| 307 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.701 |
0.287 |
| 308 |
253532_at |
AT4G31570
|
unknown |
-0.690 |
-0.138 |
| 309 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
-0.655 |
-0.068 |
| 310 |
245267_at |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.636 |
-0.022 |
| 311 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.634 |
0.300 |
| 312 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.613 |
0.931 |
| 313 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-0.596 |
0.015 |
| 314 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.581 |
0.156 |
| 315 |
258379_at |
AT3G16700
|
[Fumarylacetoacetate (FAA) hydrolase family] |
-0.538 |
0.213 |
| 316 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
-0.526 |
-0.051 |
| 317 |
246316_at |
AT3G56890
|
[F-box associated ubiquitination effector family protein] |
-0.522 |
-0.029 |
| 318 |
254582_at |
AT4G19470
|
[Leucine-rich repeat (LRR) family protein] |
-0.520 |
0.063 |
| 319 |
256210_at |
AT1G50950
|
[INVOLVED IN: cell redox homeostasis] |
-0.493 |
0.122 |
| 320 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.491 |
0.836 |
| 321 |
249454_at |
AT5G39520
|
unknown |
-0.485 |
0.603 |
| 322 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
-0.484 |
-0.159 |
| 323 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.484 |
-0.008 |
| 324 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.478 |
0.732 |
| 325 |
261001_at |
AT1G26530
|
[PIN domain-like family protein] |
-0.463 |
0.030 |
| 326 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.461 |
1.122 |
| 327 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.446 |
-0.004 |
| 328 |
259441_at |
AT1G02300
|
[Cysteine proteinases superfamily protein] |
-0.439 |
0.091 |
| 329 |
257354_x_at |
AT2G23480
|
unknown |
-0.437 |
0.005 |
| 330 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
-0.419 |
-0.001 |
| 331 |
263109_at |
AT1G65180
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.413 |
-0.066 |
| 332 |
257493_at |
AT1G48180
|
unknown |
-0.410 |
0.010 |
| 333 |
267078_at |
AT2G40960
|
[Single-stranded nucleic acid binding R3H protein] |
-0.409 |
-0.132 |
| 334 |
261463_at |
AT1G07740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.408 |
0.049 |
| 335 |
246700_at |
AT5G28030
|
DES1, L-cysteine desulfhydrase 1 |
-0.399 |
0.275 |
| 336 |
253689_at |
AT4G29770
|
unknown |
-0.398 |
-0.047 |
| 337 |
266412_at |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
-0.393 |
-0.003 |
| 338 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.390 |
0.004 |
| 339 |
254415_at |
AT4G21326
|
ATSBT3.12, subtilase 3.12, SBT3.12, subtilase 3.12 |
-0.388 |
0.089 |
| 340 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.387 |
-0.009 |
| 341 |
246394_at |
AT1G58160
|
JAX1, JACALIN-TYPE LECTIN REQUIRED FOR POTEXVIRUS RESISTANCE 1 |
-0.387 |
-0.114 |
| 342 |
260225_at |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
-0.386 |
0.795 |
| 343 |
251280_at |
AT3G61810
|
[Glycosyl hydrolase family 17 protein] |
-0.381 |
0.000 |
| 344 |
258939_at |
AT3G10020
|
unknown |
-0.379 |
0.296 |
| 345 |
261221_at |
AT1G19960
|
unknown |
-0.376 |
-0.237 |
| 346 |
259968_at |
AT1G76530
|
[Auxin efflux carrier family protein] |
-0.373 |
-0.281 |
| 347 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
-0.371 |
0.544 |
| 348 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.371 |
0.384 |
| 349 |
254948_at |
AT4G11000
|
[Ankyrin repeat family protein] |
-0.371 |
0.263 |
| 350 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.365 |
0.257 |
| 351 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
-0.364 |
-0.022 |
| 352 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.359 |
0.394 |
| 353 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-0.359 |
0.729 |
| 354 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.355 |
0.609 |
| 355 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-0.354 |
0.061 |
| 356 |
260668_at |
AT1G19530
|
unknown |
-0.350 |
0.053 |
| 357 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.348 |
0.045 |
| 358 |
245518_at |
AT4G15850
|
ATRH1, RNA helicase 1, RH1, RNA helicase 1 |
-0.347 |
-0.042 |
| 359 |
252330_at |
AT3G48770
|
[DNA binding] |
-0.346 |
-0.058 |
| 360 |
254287_at |
AT4G22960
|
unknown |
-0.343 |
0.074 |
| 361 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.337 |
0.623 |
| 362 |
266222_at |
AT2G28780
|
unknown |
-0.336 |
0.005 |
| 363 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-0.334 |
0.024 |
| 364 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
-0.332 |
0.210 |
| 365 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.330 |
-0.071 |
| 366 |
267036_at |
AT2G38465
|
unknown |
-0.329 |
0.138 |
| 367 |
262542_at |
AT1G34180
|
anac016, NAC domain containing protein 16, NAC016, NAC domain containing protein 16 |
-0.328 |
0.198 |
| 368 |
262126_at |
AT1G59620
|
CW9 |
-0.326 |
0.109 |
| 369 |
245993_at |
AT5G20700
|
unknown |
-0.324 |
-0.120 |
| 370 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
-0.324 |
0.073 |
| 371 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.324 |
0.074 |
| 372 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
-0.319 |
-0.096 |
| 373 |
262233_at |
AT1G48310
|
CHA18, CHR18, chromatin remodeling factor18 |
-0.318 |
-0.049 |
| 374 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
-0.318 |
0.087 |
| 375 |
256138_at |
AT1G48670
|
[auxin-responsive GH3 family protein] |
-0.315 |
0.018 |
| 376 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
-0.314 |
0.036 |
| 377 |
262301_at |
AT1G70880
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.314 |
-0.028 |
| 378 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.308 |
0.072 |
| 379 |
256150_at |
AT1G55120
|
AtcwINV3, 6-fructan exohydrolase, ATFRUCT5, beta-fructofuranosidase 5, FRUCT5, beta-fructofuranosidase 5 |
-0.307 |
0.025 |
| 380 |
265155_at |
AT1G30990
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.305 |
-0.110 |
| 381 |
251194_at |
AT3G62920
|
unknown |
-0.304 |
0.067 |
| 382 |
249321_at |
AT5G40920
|
[pseudogene] |
-0.300 |
-0.231 |
| 383 |
248431_at |
AT5G51470
|
[Auxin-responsive GH3 family protein] |
-0.299 |
-0.013 |
| 384 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
-0.297 |
0.518 |
| 385 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.296 |
-0.330 |
| 386 |
245560_at |
AT4G15480
|
UGT84A1 |
-0.296 |
0.032 |
| 387 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.296 |
-0.348 |
| 388 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.291 |
0.021 |
| 389 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.290 |
0.227 |
| 390 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.288 |
0.048 |
| 391 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
-0.288 |
1.239 |
| 392 |
265428_at |
AT2G20720
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.287 |
-0.037 |
| 393 |
246326_at |
AT1G16590
|
ATREV7, REV7 |
-0.284 |
0.003 |
| 394 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
-0.282 |
0.177 |
| 395 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.281 |
0.201 |
| 396 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.280 |
-0.147 |
| 397 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
-0.279 |
0.227 |
| 398 |
254377_at |
AT4G21650
|
[Subtilase family protein] |
-0.279 |
-0.169 |
| 399 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.279 |
0.005 |
| 400 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.278 |
0.048 |
| 401 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
-0.273 |
0.263 |
| 402 |
251030_at |
AT5G02130
|
NDP1 |
-0.272 |
0.023 |
| 403 |
263151_at |
AT1G54120
|
unknown |
-0.270 |
-0.090 |
| 404 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
-0.269 |
0.675 |
| 405 |
251022_at |
AT5G02150
|
Fes1C, Fes1C |
-0.269 |
0.085 |
| 406 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
-0.268 |
-0.089 |
| 407 |
262801_at |
AT1G21010
|
unknown |
-0.267 |
0.516 |
| 408 |
249495_at |
AT5G39100
|
GLP6, germin-like protein 6 |
-0.266 |
-0.020 |
| 409 |
258091_at |
AT3G14560
|
unknown |
-0.266 |
0.432 |
| 410 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
-0.265 |
0.464 |
| 411 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.265 |
0.726 |
| 412 |
253945_at |
AT4G27050
|
[F-box/RNI-like superfamily protein] |
-0.263 |
-0.140 |
| 413 |
260353_at |
AT1G69230
|
SP1L2, SPIRAL1-like2 |
-0.261 |
-0.069 |
| 414 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.259 |
-0.476 |
| 415 |
266930_at |
AT2G45930
|
unknown |
-0.252 |
0.331 |
| 416 |
256211_at |
AT1G50960
|
ATGA2OX7, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 7, GA2OX7, gibberellin 2-oxidase 7 |
-0.251 |
-0.015 |
| 417 |
262665_at |
AT1G14070
|
FUT7, fucosyltransferase 7 |
-0.251 |
0.328 |
| 418 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
-0.251 |
0.081 |
| 419 |
258348_at |
AT3G17710
|
[F-box and associated interaction domains-containing protein] |
-0.251 |
0.084 |
| 420 |
254919_at |
AT4G11360
|
RHA1B, RING-H2 finger A1B |
-0.251 |
0.007 |
| 421 |
246375_at |
AT1G51830
|
[Leucine-rich repeat protein kinase family protein] |
-0.251 |
0.017 |
| 422 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.249 |
0.386 |
| 423 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.249 |
-0.245 |
| 424 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
-0.249 |
-0.007 |
| 425 |
248990_at |
AT5G45210
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.245 |
0.035 |
| 426 |
246868_at |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-0.245 |
0.083 |
| 427 |
255262_at |
AT4G05140
|
[Nucleoside transporter family protein] |
-0.244 |
-0.062 |
| 428 |
264988_at |
AT1G27140
|
ATGSTU14, glutathione S-transferase tau 14, GST13, GLUTATHIONE S-TRANSFERASE 13, GSTU14, glutathione S-transferase tau 14 |
-0.244 |
0.088 |
| 429 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.243 |
0.347 |
| 430 |
255954_at |
AT1G22090
|
emb2204, embryo defective 2204 |
-0.242 |
-0.017 |
| 431 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
-0.241 |
-0.113 |
| 432 |
254816_at |
AT4G12440
|
APT4, adenine phosphoribosyl transferase 4 |
-0.240 |
0.023 |
| 433 |
256562_at |
AT3G29775
|
[copia-like retrotransposon family, has a 2.8e-246 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.238 |
-0.018 |
| 434 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
-0.237 |
-0.064 |
| 435 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
-0.236 |
0.168 |
| 436 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.235 |
-0.079 |
| 437 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
-0.234 |
-0.113 |
| 438 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
-0.234 |
-0.057 |
| 439 |
262421_at |
AT1G50290
|
unknown |
-0.233 |
-0.009 |
| 440 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-0.232 |
0.916 |
| 441 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.232 |
0.487 |
| 442 |
267178_at |
AT2G37750
|
unknown |
-0.231 |
0.551 |
| 443 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.230 |
-0.050 |
| 444 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.230 |
0.042 |
| 445 |
253331_at |
AT4G33490
|
[Eukaryotic aspartyl protease family protein] |
-0.229 |
0.041 |
| 446 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.229 |
0.054 |
| 447 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.229 |
0.473 |
| 448 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.228 |
-0.054 |
| 449 |
253585_at |
AT4G30720
|
PDE327, PIGMENT DEFECTIVE 327 |
-0.228 |
-0.168 |
| 450 |
257678_at |
AT3G20420
|
ATRTL2, RNASEIII-LIKE 2, RTL2, RNAse THREE-like protein 2 |
-0.227 |
-0.097 |
| 451 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
-0.227 |
0.557 |
| 452 |
254743_at |
AT4G13420
|
ATHAK5, ARABIDOPSIS THALIANA HIGH AFFINITY K+ TRANSPORTER 5, HAK5, high affinity K+ transporter 5 |
-0.226 |
0.007 |
| 453 |
252322_at |
AT3G48550
|
unknown |
-0.226 |
-0.223 |
| 454 |
261367_at |
AT1G53080
|
[Legume lectin family protein] |
-0.225 |
0.048 |
| 455 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.224 |
0.495 |
| 456 |
258890_at |
AT3G05690
|
ATHAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, HAP2B, HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B, NF-YA2, nuclear factor Y, subunit A2, UNE8, UNFERTILIZED EMBRYO SAC 8 |
-0.224 |
0.058 |
| 457 |
263104_at |
AT2G05120
|
[Nucleoporin, Nup133/Nup155-like] |
-0.224 |
-0.167 |
| 458 |
265543_at |
AT2G28270
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.224 |
-0.014 |
| 459 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
-0.222 |
0.430 |
| 460 |
252448_at |
AT3G47050
|
[Glycosyl hydrolase family protein] |
-0.220 |
-0.002 |
| 461 |
261947_at |
AT1G64470
|
[Ubiquitin-like superfamily protein] |
-0.220 |
0.230 |
| 462 |
266259_at |
AT2G27830
|
unknown |
-0.219 |
0.213 |
| 463 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.218 |
-0.071 |
| 464 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.217 |
0.163 |
| 465 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.217 |
-0.137 |
| 466 |
247250_at |
AT5G64630
|
FAS2, FASCIATA 2, MUB3.9, NFB01, NFB1 |
-0.216 |
-0.082 |
| 467 |
266234_at |
AT2G02350
|
AtPP2-B9, Phloem protein 2-B9, SKIP3, SKP1 interacting partner 3 |
-0.216 |
0.140 |
| 468 |
256923_at |
AT3G29635
|
[HXXXD-type acyl-transferase family protein] |
-0.215 |
-0.045 |
| 469 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
-0.215 |
0.685 |
| 470 |
247278_at |
AT5G64380
|
[Inositol monophosphatase family protein] |
-0.214 |
-0.211 |
| 471 |
262004_at |
AT1G64480
|
CBL8, calcineurin B-like protein 8 |
-0.214 |
0.019 |
| 472 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.213 |
0.196 |
| 473 |
248528_at |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
-0.213 |
0.491 |
| 474 |
262903_at |
AT1G59950
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.212 |
0.354 |
| 475 |
248726_at |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
-0.211 |
0.318 |
| 476 |
245873_at |
AT1G26260
|
CIB5, cryptochrome-interacting basic-helix-loop-helix 5 |
-0.209 |
-0.017 |
| 477 |
252551_at |
AT3G45880
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.209 |
0.006 |
| 478 |
247212_at |
AT5G65040
|
unknown |
-0.208 |
-0.149 |
| 479 |
262920_at |
AT1G79500
|
AtkdsA1 |
-0.208 |
-0.115 |
| 480 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.208 |
0.024 |
| 481 |
248509_at |
AT5G50335
|
unknown |
-0.208 |
0.063 |
| 482 |
244997_at |
ATCG00170
|
RPOC2 |
-0.206 |
-0.174 |
| 483 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.206 |
-0.005 |
| 484 |
256697_at |
AT3G20660
|
AtOCT4, organic cation/carnitine transporter4, OCT4, organic cation/carnitine transporter4 |
-0.206 |
0.161 |
| 485 |
262894_at |
AT1G59780
|
[NB-ARC domain-containing disease resistance protein] |
-0.206 |
0.013 |
| 486 |
256137_at |
AT1G48690
|
[Auxin-responsive GH3 family protein] |
-0.206 |
-0.056 |
| 487 |
248985_at |
AT5G45160
|
RL2, RHD-like2 |
-0.205 |
-0.048 |
| 488 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
-0.204 |
0.336 |
| 489 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.204 |
0.315 |
| 490 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.204 |
-0.061 |
| 491 |
266756_at |
AT2G46950
|
CYP709B2, cytochrome P450, family 709, subfamily B, polypeptide 2 |
-0.203 |
-0.007 |
| 492 |
252530_at |
AT3G46500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.203 |
-0.051 |
| 493 |
248189_at |
AT5G54090
|
[DNA mismatch repair protein MutS, type 2] |
-0.203 |
-0.073 |
| 494 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
-0.199 |
0.596 |
| 495 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.199 |
-0.201 |
| 496 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
-0.199 |
-0.020 |
| 497 |
255266_at |
AT4G05200
|
CRK25, cysteine-rich RLK (RECEPTOR-like protein kinase) 25 |
-0.199 |
0.001 |
| 498 |
250965_at |
AT5G03020
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.199 |
-0.023 |
| 499 |
260169_at |
AT1G71990
|
ATFT4, ARABIDOPSIS FUCOSYLTRANSFERASE 4, ATFUT13, FT4-M, FUCTC, FUT13, fucosyltransferase 13 |
-0.198 |
-0.046 |
| 500 |
258726_at |
AT3G11745
|
unknown |
-0.198 |
-0.013 |
| 501 |
247832_at |
AT5G58550
|
EOL2, ETO1-like 2 |
-0.197 |
-0.188 |
| 502 |
264314_at |
AT1G70420
|
unknown |
-0.197 |
0.387 |
| 503 |
267559_at |
AT2G45570
|
CYP76C2, cytochrome P450, family 76, subfamily C, polypeptide 2 |
-0.197 |
0.626 |
| 504 |
263572_at |
AT2G17060
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.196 |
0.043 |
| 505 |
262213_at |
AT1G74870
|
[RING/U-box superfamily protein] |
-0.196 |
-0.100 |
| 506 |
262446_at |
AT1G49310
|
unknown |
-0.195 |
0.001 |
| 507 |
258002_at |
AT3G28930
|
AIG2, AVRRPT2-INDUCED GENE 2 |
-0.195 |
0.569 |
| 508 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.194 |
-0.144 |
| 509 |
257712_at |
AT3G27420
|
unknown |
-0.193 |
0.063 |
| 510 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.193 |
0.153 |
| 511 |
267429_at |
AT2G34850
|
MEE25, maternal effect embryo arrest 25 |
-0.192 |
0.100 |
| 512 |
250732_at |
AT5G06480
|
[Immunoglobulin E-set superfamily protein] |
-0.192 |
-0.026 |
| 513 |
259535_at |
AT1G12280
|
SUMM2, suppressor of mkk1 mkk2 2 |
-0.191 |
-0.373 |
| 514 |
251576_at |
AT3G58200
|
[TRAF-like family protein] |
-0.191 |
-0.144 |
| 515 |
262452_at |
AT1G11210
|
unknown |
-0.190 |
0.762 |
| 516 |
262132_at |
AT1G02830
|
[Ribosomal L22e protein family] |
-0.190 |
0.023 |
| 517 |
259876_at |
AT1G76700
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.189 |
0.133 |
| 518 |
261636_at |
AT1G50110
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.189 |
-0.307 |
| 519 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.189 |
-0.007 |
| 520 |
245692_at |
AT5G04150
|
BHLH101 |
-0.189 |
-0.605 |
| 521 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.188 |
0.271 |
| 522 |
260684_at |
AT1G17590
|
NF-YA8, nuclear factor Y, subunit A8 |
-0.188 |
0.037 |
| 523 |
267207_at |
AT2G30840
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.188 |
0.183 |
| 524 |
245608_at |
AT4G14350
|
[AGC (cAMP-dependent, cGMP-dependent and protein kinase C) kinase family protein] |
-0.188 |
-0.000 |
| 525 |
254496_at |
AT4G20070
|
AAH, allantoate amidohydrolase, ATAAH, allantoate amidohydrolase |
-0.187 |
-0.232 |
| 526 |
247848_at |
AT5G58120
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.187 |
-0.045 |
| 527 |
261883_at |
AT1G80870
|
[Protein kinase superfamily protein] |
-0.187 |
-0.258 |
| 528 |
246731_at |
AT5G27630
|
ACBP5, acyl-CoA binding protein 5, AtACBP5 |
-0.187 |
-0.048 |
| 529 |
245602_at |
AT4G14270
|
unknown |
-0.187 |
0.151 |
| 530 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.186 |
-0.588 |
| 531 |
250828_at |
AT5G05250
|
unknown |
-0.186 |
-0.367 |
| 532 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.186 |
-0.671 |
| 533 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.186 |
-0.448 |
| 534 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
-0.186 |
-0.008 |
| 535 |
250028_at |
AT5G18130
|
unknown |
-0.185 |
0.388 |
| 536 |
249724_at |
AT5G35450
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.184 |
-0.362 |
| 537 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.183 |
-0.244 |
| 538 |
262888_at |
AT1G14790
|
AtRDR1, ATRDRP1, RDR1, RNA-dependent RNA polymerase 1 |
-0.183 |
-0.059 |
| 539 |
247015_at |
AT5G66960
|
[Prolyl oligopeptidase family protein] |
-0.182 |
-0.107 |
| 540 |
261894_at |
AT1G80900
|
ATMGT1, MAGNESIUM TRANSPORTER 1, MGT1, magnesium transporter 1, MRS2-10 |
-0.181 |
0.006 |
| 541 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.181 |
-0.044 |
| 542 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.180 |
-0.190 |
| 543 |
255013_at |
AT4G10000
|
[Thioredoxin family protein] |
-0.180 |
0.029 |
| 544 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.179 |
-0.036 |
| 545 |
253911_at |
AT4G27300
|
[S-locus lectin protein kinase family protein] |
-0.179 |
0.083 |
| 546 |
266988_at |
AT2G39310
|
JAL22, jacalin-related lectin 22 |
-0.179 |
0.414 |
| 547 |
254781_at |
AT4G12840
|
unknown |
-0.179 |
0.050 |
| 548 |
266308_at |
AT2G27010
|
CYP705A9, cytochrome P450, family 705, subfamily A, polypeptide 9 |
-0.178 |
-0.001 |
| 549 |
245723_at |
AT1G73400
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.178 |
-0.039 |
| 550 |
264271_at |
AT1G60270
|
BGLU6, beta glucosidase 6 |
-0.178 |
-0.004 |
| 551 |
249358_at |
AT5G40510
|
[Sucrase/ferredoxin-like family protein] |
-0.178 |
0.103 |
| 552 |
246055_at |
AT5G08380
|
AGAL1, alpha-galactosidase 1, AtAGAL1, alpha-galactosidase 1 |
-0.177 |
-0.002 |
| 553 |
265584_at |
AT2G20180
|
PIF1, PHY-INTERACTING FACTOR 1, PIL5, phytochrome interacting factor 3-like 5 |
-0.177 |
-0.488 |
| 554 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
-0.177 |
0.090 |
| 555 |
249422_at |
AT5G39760
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23, ZHD10, ZINC FINGER HOMEODOMAIN 10 |
-0.176 |
0.168 |
| 556 |
254612_at |
AT4G19100
|
PAM68, photosynthesis affected mutant 68 |
-0.176 |
-0.169 |
| 557 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.176 |
0.327 |
| 558 |
259544_at |
AT1G20620
|
ATCAT3, CAT3, catalase 3, SEN2, SENESCENCE 2 |
-0.176 |
0.100 |
| 559 |
263546_at |
AT2G21550
|
[Bifunctional dihydrofolate reductase/thymidylate synthase] |
-0.176 |
0.021 |
| 560 |
257154_at |
AT3G27210
|
unknown |
-0.176 |
0.793 |
| 561 |
249309_at |
AT5G41410
|
BEL1, BELL 1 |
-0.175 |
-0.094 |
| 562 |
261708_at |
AT1G32740
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.175 |
0.217 |
| 563 |
253358_at |
AT4G32940
|
GAMMA-VPE, gamma vacuolar processing enzyme, GAMMAVPE |
-0.175 |
0.064 |
| 564 |
259624_at |
AT1G43020
|
unknown |
-0.174 |
-0.007 |
| 565 |
254797_at |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.174 |
0.168 |
| 566 |
263176_at |
AT1G05530
|
UGT2, UDP-GLUCOSYL TRANSFERASE 2, UGT75B2, UDP-glucosyl transferase 75B2 |
-0.174 |
-0.042 |
| 567 |
263174_at |
AT1G54040
|
ESP, epithiospecifier protein, ESR, EPITHIOSPECIFYING SENESCENCE REGULATOR, TASTY |
-0.172 |
0.123 |
| 568 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.172 |
-0.257 |
| 569 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.172 |
-0.027 |
| 570 |
260843_at |
AT1G29060
|
[Target SNARE coiled-coil domain protein] |
-0.172 |
0.182 |
| 571 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.171 |
-0.112 |
| 572 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.171 |
-0.019 |
| 573 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
-0.171 |
0.140 |
| 574 |
255133_at |
AT4G08320
|
TPR8, tetratricopeptide repeat 8 |
-0.170 |
0.057 |
| 575 |
249286_at |
AT5G41580
|
[RING/U-box superfamily protein] |
-0.169 |
0.170 |
| 576 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
-0.168 |
0.043 |
| 577 |
247118_at |
AT5G65890
|
ACR1, ACT domain repeat 1 |
-0.168 |
-0.007 |
| 578 |
263897_at |
AT2G21940
|
ATSK1, SK1, shikimate kinase 1 |
-0.167 |
-0.098 |
| 579 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.167 |
-0.227 |
| 580 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.166 |
-0.209 |
| 581 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
-0.166 |
-0.086 |
| 582 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
-0.166 |
0.650 |
| 583 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.166 |
0.043 |
| 584 |
261369_at |
AT1G53060
|
[Legume lectin family protein] |
-0.166 |
0.053 |
| 585 |
266529_at |
AT2G16880
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.165 |
-0.034 |
| 586 |
249601_at |
AT5G37980
|
[Zinc-binding dehydrogenase family protein] |
-0.165 |
0.011 |
| 587 |
258042_at |
AT3G21310
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.164 |
-0.025 |
| 588 |
265276_at |
AT2G28400
|
unknown |
-0.164 |
0.875 |
| 589 |
247800_at |
AT5G58570
|
unknown |
-0.164 |
-0.044 |
| 590 |
259534_at |
AT1G12290
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.163 |
0.128 |
| 591 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.163 |
0.278 |
| 592 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.163 |
0.207 |
| 593 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.163 |
0.013 |
| 594 |
264161_at |
AT1G65420
|
NPQ7, NONPHOTOCHEMICAL QUENCHING 7 |
-0.162 |
-0.054 |
| 595 |
247528_at |
AT5G61460
|
ATRAD18, MIM, hypersensitive to MMS, irradiation and MMC, SMC6B, STRUCTURAL MAINTENANCE OF CHROMOSOMES 6B |
-0.161 |
-0.075 |
| 596 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.161 |
-0.643 |
| 597 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.161 |
0.053 |
| 598 |
265197_at |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-0.161 |
0.003 |
| 599 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
-0.161 |
0.351 |
| 600 |
256066_at |
AT1G06980
|
unknown |
-0.160 |
-0.405 |