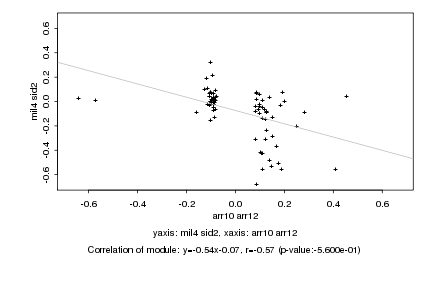
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
arr10 arr12 |
Log2 signal ratio
mil4 sid2 |
| 1 |
245532_at |
AT4G15110
|
CYP97B3, cytochrome P450, family 97, subfamily B, polypeptide 3 |
0.453 |
0.046 |
| 2 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.407 |
-0.550 |
| 3 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.282 |
-0.085 |
| 4 |
256603_at |
AT3G28270
|
unknown |
0.247 |
-0.201 |
| 5 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.200 |
0.008 |
| 6 |
262719_at |
AT1G43590
|
unknown |
0.192 |
0.082 |
| 7 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.186 |
-0.553 |
| 8 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.181 |
-0.025 |
| 9 |
264102_at |
AT1G79270
|
ECT8, evolutionarily conserved C-terminal region 8 |
0.172 |
-0.507 |
| 10 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.166 |
-0.369 |
| 11 |
259760_at |
AT1G77580
|
unknown |
0.150 |
-0.125 |
| 12 |
267166_at |
AT2G37720
|
TBL15, TRICHOME BIREFRINGENCE-LIKE 15 |
0.150 |
-0.279 |
| 13 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.145 |
-0.531 |
| 14 |
254716_at |
AT4G13560
|
UNE15, unfertilized embryo sac 15 |
0.139 |
0.035 |
| 15 |
263595_at |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
0.136 |
-0.481 |
| 16 |
261922_at |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
0.126 |
-0.086 |
| 17 |
256924_at |
AT3G29590
|
AT5MAT |
0.124 |
-0.077 |
| 18 |
248205_at |
AT5G54300
|
unknown |
0.123 |
-0.085 |
| 19 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.123 |
-0.235 |
| 20 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
0.120 |
-0.147 |
| 21 |
258982_at |
AT3G08870
|
LecRK-VI.1, L-type lectin receptor kinase VI.1 |
0.119 |
-0.309 |
| 22 |
250479_at |
AT5G10260
|
AtRABH1e, RAB GTPase homolog H1E, RABH1e, RAB GTPase homolog H1E |
0.118 |
-0.059 |
| 23 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.110 |
-0.042 |
| 24 |
256093_at |
AT1G20823
|
[RING/U-box superfamily protein] |
0.109 |
-0.426 |
| 25 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.108 |
0.009 |
| 26 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.108 |
-0.551 |
| 27 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
0.108 |
-0.134 |
| 28 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
0.105 |
-0.425 |
| 29 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.100 |
-0.412 |
| 30 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.098 |
-0.098 |
| 31 |
256540_at |
AT3G32917
|
[gypsy-like retrotransposon family, has a 1.0e-114 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
0.097 |
-0.024 |
| 32 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.095 |
0.059 |
| 33 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
0.095 |
-0.034 |
| 34 |
249843_at |
AT5G23570
|
ATSGS3, SUPPRESSOR OF GENE SILENCING 3, SGS3, SUPPRESSOR OF GENE SILENCING 3 |
0.094 |
-0.063 |
| 35 |
250503_at |
AT5G09820
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
0.085 |
0.021 |
| 36 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.083 |
-0.673 |
| 37 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
0.083 |
0.078 |
| 38 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.082 |
0.073 |
| 39 |
262751_at |
AT1G16310
|
[Cation efflux family protein] |
0.081 |
-0.037 |
| 40 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
0.080 |
-0.304 |
| 41 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
0.080 |
-0.075 |
| 42 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.644 |
0.030 |
| 43 |
264374_at |
AT2G25180
|
ARR12, response regulator 12, RR12, response regulator 12 |
-0.574 |
0.009 |
| 44 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.162 |
-0.084 |
| 45 |
263901_at |
AT2G36320
|
[A20/AN1-like zinc finger family protein] |
-0.127 |
0.104 |
| 46 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.122 |
0.197 |
| 47 |
261924_at |
AT1G22550
|
[Major facilitator superfamily protein] |
-0.118 |
0.110 |
| 48 |
256709_at |
AT3G30330
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.7e-83 P-value blast match to At1g36190.1/92-340 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.115 |
-0.017 |
| 49 |
250264_at |
AT5G12890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.110 |
0.069 |
| 50 |
261472_at |
AT1G14470
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.108 |
0.045 |
| 51 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.108 |
0.043 |
| 52 |
265839_at |
AT2G14540
|
ATSRP2, SRP2, serpin 2 |
-0.107 |
-0.019 |
| 53 |
261807_at |
AT1G30515
|
unknown |
-0.107 |
-0.025 |
| 54 |
255219_at |
AT4G07720
|
[pseudogene] |
-0.106 |
0.001 |
| 55 |
249479_at |
AT5G38960
|
[RmlC-like cupins superfamily protein] |
-0.106 |
0.081 |
| 56 |
250157_at |
AT5G15180
|
[Peroxidase superfamily protein] |
-0.106 |
0.078 |
| 57 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.103 |
0.321 |
| 58 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-0.102 |
-0.150 |
| 59 |
251748_at |
AT3G55680
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.102 |
0.004 |
| 60 |
252330_at |
AT3G48770
|
[DNA binding] |
-0.101 |
0.020 |
| 61 |
247344_at |
AT5G63750
|
ARI13, ARIADNE 13, ATARI13, ARABIDOPSIS ARIADNE 13 |
-0.100 |
0.066 |
| 62 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.098 |
-0.001 |
| 63 |
261690_at |
AT1G50090
|
AtBCAT7, BCAT7, branched-chain amino acid transaminase 7 |
-0.094 |
0.019 |
| 64 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-0.094 |
0.219 |
| 65 |
247016_at |
AT5G66970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.093 |
0.014 |
| 66 |
262027_at |
AT1G35515
|
HOS10, high response to osmotic stress 10, MYB8 |
-0.093 |
-0.018 |
| 67 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
-0.093 |
-0.049 |
| 68 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.092 |
0.072 |
| 69 |
250309_at |
AT5G12220
|
[las1-like family protein] |
-0.092 |
-0.020 |
| 70 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
-0.091 |
-0.073 |
| 71 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-0.091 |
0.039 |
| 72 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.091 |
0.041 |
| 73 |
267042_at |
AT2G34320
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.089 |
0.018 |
| 74 |
255831_at |
AT2G33350
|
[CCT motif family protein] |
-0.089 |
0.009 |
| 75 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
-0.088 |
-0.129 |
| 76 |
247369_at |
AT5G63340
|
unknown |
-0.087 |
-0.000 |
| 77 |
249012_at |
AT5G44620
|
CYP706A3, cytochrome P450, family 706, subfamily A, polypeptide 3 |
-0.084 |
0.040 |
| 78 |
258600_at |
AT3G02810
|
LIP2, LOST IN POLLEN TUBE GUIDANCE 2 |
-0.084 |
0.015 |
| 79 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.083 |
-0.062 |
| 80 |
247731_at |
AT5G59590
|
UGT76E2, UDP-glucosyl transferase 76E2 |
-0.083 |
0.098 |
| 81 |
251284_at |
AT3G61840
|
unknown |
-0.081 |
0.043 |