|
probeID |
AGICode |
Annotation |
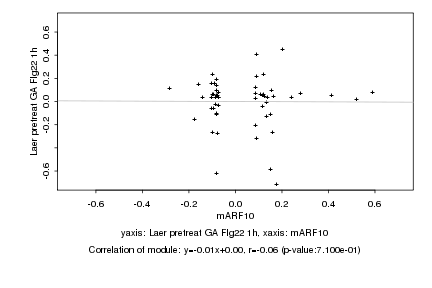
Log2 signal ratio
mARF10 |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
262431_at |
AT1G47540
|
[Scorpion toxin-like knottin superfamily protein] |
0.587 |
0.079 |
| 2 |
253904_at |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
0.517 |
0.024 |
| 3 |
260458_at |
AT1G68250
|
unknown |
0.409 |
0.058 |
| 4 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
0.277 |
0.070 |
| 5 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
0.239 |
0.040 |
| 6 |
251612_at |
AT3G57950
|
unknown |
0.202 |
0.454 |
| 7 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.176 |
-0.708 |
| 8 |
258178_at |
AT3G21680
|
unknown |
0.160 |
0.051 |
| 9 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
0.157 |
-0.267 |
| 10 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
0.151 |
0.099 |
| 11 |
261247_at |
AT1G20070
|
unknown |
0.148 |
-0.112 |
| 12 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.148 |
-0.584 |
| 13 |
247190_at |
AT5G65420
|
CYCD4;1, CYCLIN D4;1 |
0.134 |
0.039 |
| 14 |
255427_at |
AT4G03380
|
unknown |
0.131 |
-0.001 |
| 15 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.130 |
-0.129 |
| 16 |
261369_at |
AT1G53060
|
[Legume lectin family protein] |
0.121 |
0.049 |
| 17 |
265382_at |
AT2G16790
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.120 |
0.237 |
| 18 |
260925_at |
AT1G21340
|
[Dof-type zinc finger DNA-binding family protein] |
0.117 |
0.061 |
| 19 |
248642_at |
AT5G49120
|
unknown |
0.115 |
0.054 |
| 20 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.114 |
-0.038 |
| 21 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
0.107 |
0.067 |
| 22 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.090 |
0.410 |
| 23 |
249916_at |
AT5G22880
|
H2B, HISTONE H2B, HTB2, histone B2 |
0.087 |
0.217 |
| 24 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.087 |
-0.316 |
| 25 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.084 |
-0.205 |
| 26 |
262622_at |
AT1G06510
|
unknown |
0.083 |
0.126 |
| 27 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
0.083 |
0.033 |
| 28 |
261923_at |
AT1G22380
|
AtUGT85A3, UDP-glucosyl transferase 85A3, UGT85A3, UDP-glucosyl transferase 85A3 |
0.082 |
0.077 |
| 29 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.286 |
0.120 |
| 30 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.178 |
-0.151 |
| 31 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.159 |
0.147 |
| 32 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.145 |
0.037 |
| 33 |
251749_at |
AT3G55690
|
unknown |
-0.105 |
0.042 |
| 34 |
254515_at |
AT4G20270
|
BAM3, BARELY ANY MERISTEM 3 |
-0.103 |
-0.053 |
| 35 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
-0.103 |
0.160 |
| 36 |
258756_at |
AT3G11960
|
[Cleavage and polyadenylation specificity factor (CPSF) A subunit protein] |
-0.102 |
0.064 |
| 37 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.099 |
-0.262 |
| 38 |
251011_at |
AT5G02560
|
HTA12, histone H2A 12 |
-0.099 |
0.234 |
| 39 |
251050_at |
AT5G02440
|
unknown |
-0.098 |
0.065 |
| 40 |
256220_at |
AT1G56230
|
unknown |
-0.097 |
-0.052 |
| 41 |
257829_at |
AT3G26680
|
ATSNM1, SENSITIVE TO NITROGEN MUSTARD 1, SNM1, SENSITIVE TO NITROGEN MUSTARD 1 |
-0.092 |
0.156 |
| 42 |
260924_at |
AT1G21590
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.090 |
0.039 |
| 43 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.089 |
-0.022 |
| 44 |
264539_at |
AT1G55590
|
[RNI-like superfamily protein] |
-0.084 |
0.143 |
| 45 |
265060_at |
AT1G52150
|
ATHB-15, ATHB15, CNA, CORONA, ICU4, INCURVATA 4 |
-0.083 |
-0.106 |
| 46 |
255414_at |
AT4G03156
|
[small GTPase-related] |
-0.083 |
0.058 |
| 47 |
260054_at |
AT1G78130
|
UNE2, unfertilized embryo sac 2 |
-0.083 |
0.194 |
| 48 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.083 |
-0.621 |
| 49 |
259994_at |
AT1G68130
|
AtIDD14, indeterminate(ID)-domain 14, IDD14, indeterminate(ID)-domain 14, IDD14alpha, IDD14beta |
-0.082 |
-0.101 |
| 50 |
247591_at |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-0.082 |
0.101 |
| 51 |
247511_at |
AT5G62040
|
BFT, brother of FT and TFL1 |
-0.078 |
0.055 |
| 52 |
258460_at |
AT3G17330
|
ECT6, evolutionarily conserved C-terminal region 6 |
-0.078 |
-0.274 |
| 53 |
254128_at |
AT4G24560
|
UBP16, ubiquitin-specific protease 16 |
-0.077 |
0.042 |
| 54 |
264060_at |
AT2G27980
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
-0.076 |
0.080 |
| 55 |
265591_at |
AT2G20150
|
unknown |
-0.075 |
0.040 |
| 56 |
256198_at |
AT1G58220
|
[Homeodomain-like superfamily protein] |
-0.075 |
-0.028 |