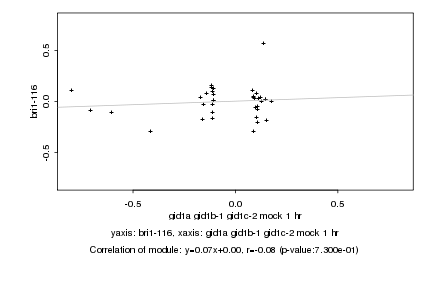
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
gid1a gid1b-1 gid1c-2 mock 1 hr |
Log2 signal ratio
bri1-116 |
| 1 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.176 |
0.007 |
| 2 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.147 |
-0.186 |
| 3 |
249696_at |
AT5G35820
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.144 |
0.020 |
| 4 |
263769_at |
AT2G06390
|
transposable element gene |
0.143 |
0.022 |
| 5 |
247800_at |
AT5G58570
|
unknown |
0.133 |
0.577 |
| 6 |
266084_at |
AT2G37810
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.127 |
0.002 |
| 7 |
250989_at |
AT5G02900
|
CYP96A13, cytochrome P450, family 96, subfamily A, polypeptide 13 |
0.122 |
0.041 |
| 8 |
260721_at |
AT1G48060
|
[F-box and associated interaction domains-containing protein] |
0.110 |
0.035 |
| 9 |
253047_at |
AT4G37295
|
unknown |
0.107 |
-0.069 |
| 10 |
251103_at |
AT5G01700
|
[Protein phosphatase 2C family protein] |
0.105 |
-0.198 |
| 11 |
245963_at |
AT5G19710
|
unknown |
0.104 |
-0.042 |
| 12 |
257514_at |
AT1G43940
|
unknown |
0.102 |
0.088 |
| 13 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.098 |
-0.153 |
| 14 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
0.095 |
-0.053 |
| 15 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.092 |
0.036 |
| 16 |
250951_at |
AT5G03550
|
unknown |
0.087 |
-0.289 |
| 17 |
255146_at |
AT4G08450
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.087 |
0.041 |
| 18 |
251987_at |
AT3G53280
|
CYP71B5, cytochrome p450 71b5 |
0.084 |
0.050 |
| 19 |
252951_at |
AT4G38700
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.083 |
0.115 |
| 20 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
-0.804 |
0.109 |
| 21 |
255461_at |
AT4G02780
|
ABC33, ATCPS1, ARABIDOPSIS THALIANA ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, CPS, CPP synthase, CPS1, ENT-COPALYL DIPHOSPHATE SYNTHETASE 1, GA1, GA REQUIRING 1 |
-0.710 |
-0.086 |
| 22 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
-0.610 |
-0.101 |
| 23 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
-0.418 |
-0.287 |
| 24 |
250387_at |
AT5G11290
|
unknown |
-0.172 |
0.044 |
| 25 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.164 |
-0.170 |
| 26 |
260669_at |
AT1G19340
|
[Methyltransferase MT-A70 family protein] |
-0.160 |
-0.022 |
| 27 |
259691_at |
AT1G63200
|
[Cystatin/monellin superfamily protein] |
-0.143 |
0.084 |
| 28 |
248650_at |
AT5G49250
|
[Beta-galactosidase related protein] |
-0.118 |
0.146 |
| 29 |
263784_at |
AT2G46375
|
unknown |
-0.118 |
0.161 |
| 30 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
-0.115 |
-0.107 |
| 31 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.115 |
-0.023 |
| 32 |
266976_at |
AT2G39410
|
[alpha/beta-Hydrolases superfamily protein] |
-0.114 |
0.108 |
| 33 |
264643_at |
AT1G08990
|
GUX5, Glucuronic Acid Substitution of Xylan 5, PGSIP5, plant glycogenin-like starch initiation protein 5 |
-0.113 |
-0.162 |
| 34 |
248872_at |
AT5G46795
|
MSP2, microspore-specific promoter 2 |
-0.111 |
0.013 |
| 35 |
248931_at |
AT5G46040
|
[Major facilitator superfamily protein] |
-0.110 |
0.019 |
| 36 |
263977_at |
AT2G42660
|
[Homeodomain-like superfamily protein] |
-0.108 |
0.078 |
| 37 |
258220_at |
AT3G17830
|
DJA4, DNA J protein A4 |
-0.108 |
0.133 |