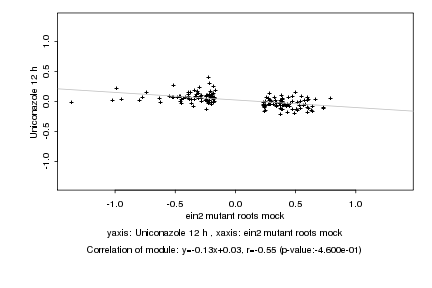
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ein2 mutant roots mock |
Log2 signal ratio
Uniconazole 12 h |
| 1 |
260582_at |
AT2G47200
|
unknown |
0.782 |
0.055 |
| 2 |
249686_at |
AT5G36140
|
CYP716A2, cytochrome P450, family 716, subfamily A, polypeptide 2 |
0.727 |
-0.099 |
| 3 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
0.724 |
-0.106 |
| 4 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.665 |
0.046 |
| 5 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.638 |
-0.150 |
| 6 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.637 |
-0.078 |
| 7 |
248621_at |
AT5G49350
|
[Glycine-rich protein family] |
0.630 |
-0.144 |
| 8 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.601 |
0.042 |
| 9 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.599 |
-0.112 |
| 10 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.598 |
0.020 |
| 11 |
257428_at |
AT1G78990
|
[HXXXD-type acyl-transferase family protein] |
0.597 |
-0.179 |
| 12 |
260438_at |
AT1G68290
|
ENDO2, endonuclease 2 |
0.594 |
0.077 |
| 13 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
0.593 |
-0.095 |
| 14 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
0.578 |
-0.034 |
| 15 |
249687_at |
AT5G36150
|
ATPEN3, putative pentacyclic triterpene synthase 3, PEN3, putative pentacyclic triterpene synthase 3 |
0.573 |
0.032 |
| 16 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.561 |
-0.063 |
| 17 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.544 |
0.084 |
| 18 |
250576_at |
AT5G08250
|
[Cytochrome P450 superfamily protein] |
0.537 |
0.003 |
| 19 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.536 |
-0.105 |
| 20 |
254474_at |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
0.527 |
-0.057 |
| 21 |
260535_at |
AT2G43390
|
unknown |
0.511 |
-0.013 |
| 22 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
0.508 |
-0.147 |
| 23 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.504 |
-0.117 |
| 24 |
265083_at |
AT1G03820
|
unknown |
0.501 |
-0.118 |
| 25 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.498 |
0.157 |
| 26 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.487 |
-0.191 |
| 27 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.469 |
0.096 |
| 28 |
257718_at |
AT3G18400
|
anac058, NAC domain containing protein 58, NAC058, NAC domain containing protein 58 |
0.467 |
0.002 |
| 29 |
246067_at |
AT5G19410
|
ABCG23, ATP-binding cassette G23 |
0.466 |
-0.133 |
| 30 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.451 |
-0.024 |
| 31 |
266006_at |
AT2G37360
|
ABCG2, ATP-binding cassette G2 |
0.449 |
-0.078 |
| 32 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
0.435 |
0.068 |
| 33 |
262406_at |
AT1G34670
|
AtMYB93, myb domain protein 93, MYB93, myb domain protein 93 |
0.431 |
-0.178 |
| 34 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.431 |
-0.055 |
| 35 |
258625_at |
AT3G04370
|
PDLP4, plasmodesmata-located protein 4 |
0.431 |
-0.041 |
| 36 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.424 |
-0.074 |
| 37 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
0.423 |
-0.076 |
| 38 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.414 |
-0.061 |
| 39 |
256217_at |
AT1G56320
|
unknown |
0.412 |
-0.053 |
| 40 |
265014_at |
AT1G24430
|
[HXXXD-type acyl-transferase family protein] |
0.403 |
-0.046 |
| 41 |
264635_at |
AT1G65500
|
unknown |
0.394 |
-0.054 |
| 42 |
252395_at |
AT3G47950
|
AHA4, H(+)-ATPase 4, HA4, H(+)-ATPase 4 |
0.390 |
0.050 |
| 43 |
264842_at |
AT1G03700
|
[Uncharacterised protein family (UPF0497)] |
0.388 |
-0.055 |
| 44 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.387 |
0.054 |
| 45 |
258905_at |
AT3G06390
|
[Uncharacterised protein family (UPF0497)] |
0.387 |
0.010 |
| 46 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.384 |
-0.122 |
| 47 |
263650_at |
AT1G04360
|
[RING/U-box superfamily protein] |
0.376 |
-0.003 |
| 48 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.376 |
0.101 |
| 49 |
262630_at |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
0.375 |
-0.125 |
| 50 |
255690_at |
AT4G00360
|
ATT1, ABERRANT INDUCTION OF TYPE THREE 1, CYP86A2, cytochrome P450, family 86, subfamily A, polypeptide 2 |
0.374 |
-0.203 |
| 51 |
267344_at |
AT2G44230
|
unknown |
0.369 |
-0.106 |
| 52 |
248761_at |
AT5G47635
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.367 |
0.032 |
| 53 |
261545_at |
AT1G63530
|
unknown |
0.367 |
-0.051 |
| 54 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.365 |
-0.022 |
| 55 |
245612_at |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
0.341 |
-0.000 |
| 56 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.340 |
-0.063 |
| 57 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.339 |
-0.065 |
| 58 |
266708_at |
AT2G03200
|
[Eukaryotic aspartyl protease family protein] |
0.334 |
-0.077 |
| 59 |
262312_at |
AT1G70830
|
MLP28, MLP-like protein 28 |
0.328 |
0.019 |
| 60 |
253195_at |
AT4G35420
|
DRL1, dihydroflavonol 4-reductase-like1, TKPR1, tetraketide alpha-pyrone reductase 1 |
0.326 |
-0.016 |
| 61 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
0.322 |
0.078 |
| 62 |
266597_at |
AT2G46130
|
ATWRKY43, WRKY DNA-BINDING PROTEIN 43, WRKY43, WRKY DNA-binding protein 43 |
0.315 |
-0.045 |
| 63 |
259735_at |
AT1G64405
|
unknown |
0.296 |
0.008 |
| 64 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.287 |
0.027 |
| 65 |
265685_at |
AT2G24430
|
ANAC038, NAC domain containing protein 38, ANAC039, Arabidopsis NAC domain containing protein 39, NAC038, NAC domain containing protein 38 |
0.286 |
-0.041 |
| 66 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.282 |
0.148 |
| 67 |
251481_at |
AT3G59730
|
LecRK-V.6, L-type lectin receptor kinase V.6 |
0.280 |
-0.034 |
| 68 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
0.277 |
0.024 |
| 69 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
0.274 |
0.063 |
| 70 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.268 |
-0.037 |
| 71 |
257919_at |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
0.253 |
0.081 |
| 72 |
256128_at |
AT1G18140
|
ATLAC1, LAC1, laccase 1 |
0.246 |
-0.053 |
| 73 |
252905_at |
AT4G39720
|
[VQ motif-containing protein] |
0.245 |
-0.136 |
| 74 |
258620_at |
AT3G02940
|
AtMYB107, myb domain protein 107, MYB107, myb domain protein 107 |
0.245 |
0.011 |
| 75 |
247769_at |
AT5G58910
|
LAC16, laccase 16 |
0.244 |
-0.053 |
| 76 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.240 |
-0.067 |
| 77 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.239 |
-0.162 |
| 78 |
257512_at |
AT1G35250
|
ALT2, acyl-lipid thioesterase 2 |
0.237 |
-0.085 |
| 79 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.237 |
-0.081 |
| 80 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
0.237 |
-0.057 |
| 81 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
0.233 |
-0.016 |
| 82 |
248100_at |
AT5G55180
|
[O-Glycosyl hydrolases family 17 protein] |
0.231 |
-0.055 |
| 83 |
263010_at |
AT1G23330
|
[alpha/beta-Hydrolases superfamily protein] |
0.228 |
-0.034 |
| 84 |
254805_at |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-1.368 |
-0.003 |
| 85 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-1.023 |
0.023 |
| 86 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
-0.994 |
0.224 |
| 87 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.949 |
0.043 |
| 88 |
251974_at |
AT3G53200
|
AtMYB27, myb domain protein 27, MYB27, myb domain protein 27 |
-0.800 |
0.022 |
| 89 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
-0.779 |
0.076 |
| 90 |
261606_at |
AT1G49570
|
[Peroxidase superfamily protein] |
-0.747 |
0.159 |
| 91 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.634 |
0.053 |
| 92 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.626 |
-0.015 |
| 93 |
258923_at |
AT3G10450
|
SCPL7, serine carboxypeptidase-like 7 |
-0.555 |
0.094 |
| 94 |
247679_at |
AT5G59540
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.525 |
0.081 |
| 95 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
-0.516 |
0.079 |
| 96 |
260668_at |
AT1G19530
|
unknown |
-0.515 |
0.272 |
| 97 |
250152_at |
AT5G15120
|
unknown |
-0.489 |
0.074 |
| 98 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.472 |
0.087 |
| 99 |
249384_at |
AT5G39890
|
unknown |
-0.461 |
0.017 |
| 100 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.458 |
0.094 |
| 101 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.450 |
-0.023 |
| 102 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.449 |
0.035 |
| 103 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.439 |
0.057 |
| 104 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.422 |
0.078 |
| 105 |
266900_at |
AT2G34610
|
unknown |
-0.396 |
0.052 |
| 106 |
261567_at |
AT1G33055
|
unknown |
-0.394 |
0.095 |
| 107 |
258570_at |
AT3G04530
|
ATPPCK2, PEPCK2, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 2, PPCK2, phosphoenolpyruvate carboxylase kinase 2 |
-0.392 |
0.133 |
| 108 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.391 |
0.152 |
| 109 |
263436_at |
AT2G28690
|
unknown |
-0.387 |
0.045 |
| 110 |
265481_at |
AT2G15960
|
unknown |
-0.377 |
0.165 |
| 111 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.367 |
0.034 |
| 112 |
262638_at |
AT1G06650
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.366 |
-0.017 |
| 113 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.351 |
-0.067 |
| 114 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.347 |
0.189 |
| 115 |
250928_at |
AT5G03280
|
ATEIN2, CKR1, CYTOKININ RESISTANT 1, EIN2, ETHYLENE INSENSITIVE 2, ERA3, ENHANCED RESPONSE TO ABA3, ORE2, ORESARA 2, ORE3, ORESARA 3, PIR2 |
-0.344 |
0.048 |
| 116 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.340 |
0.089 |
| 117 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.329 |
0.130 |
| 118 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.327 |
0.085 |
| 119 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.323 |
0.174 |
| 120 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
-0.313 |
0.138 |
| 121 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.308 |
0.054 |
| 122 |
262373_at |
AT1G73120
|
unknown |
-0.301 |
0.240 |
| 123 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
-0.298 |
0.084 |
| 124 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
-0.287 |
0.054 |
| 125 |
257026_at |
AT3G19200
|
unknown |
-0.285 |
0.002 |
| 126 |
264425_at |
AT1G61750
|
[Receptor-like protein kinase-related family protein] |
-0.282 |
0.115 |
| 127 |
252282_at |
AT3G49360
|
PGL2, 6-phosphogluconolactonase 2 |
-0.254 |
0.033 |
| 128 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
-0.247 |
0.095 |
| 129 |
248432_at |
AT5G51390
|
unknown |
-0.244 |
0.104 |
| 130 |
254550_at |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-0.244 |
-0.129 |
| 131 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-0.243 |
0.100 |
| 132 |
266757_at |
AT2G46940
|
unknown |
-0.242 |
0.042 |
| 133 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.237 |
0.013 |
| 134 |
251066_at |
AT5G01880
|
DAFL2, DAF-Like gene 2 |
-0.234 |
0.115 |
| 135 |
256681_at |
AT3G52340
|
ATSPP2, SUCROSE-PHOSPHATASE 2, SPP2, sucrose-6F-phosphate phosphohydrolase 2 |
-0.232 |
-0.023 |
| 136 |
250952_at |
AT5G03570
|
ATIREG2, ARABIDOPSIS THALIANA IRON-REGULATED PROTEIN 2, FPN2, FERROPORTIN 2, IREG2, iron regulated 2 |
-0.230 |
0.000 |
| 137 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.227 |
0.416 |
| 138 |
259982_at |
AT1G76410
|
ATL8 |
-0.223 |
0.302 |
| 139 |
247793_at |
AT5G58650
|
PSY1, plant peptide containing sulfated tyrosine 1 |
-0.223 |
0.129 |
| 140 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-0.220 |
0.119 |
| 141 |
256923_at |
AT3G29635
|
[HXXXD-type acyl-transferase family protein] |
-0.217 |
0.018 |
| 142 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.214 |
0.076 |
| 143 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.213 |
0.095 |
| 144 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-0.209 |
0.169 |
| 145 |
250158_at |
AT5G15190
|
unknown |
-0.206 |
-0.014 |
| 146 |
266444_at |
AT2G43150
|
[Proline-rich extensin-like family protein] |
-0.202 |
-0.049 |
| 147 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
-0.200 |
0.120 |
| 148 |
250937_at |
AT5G03230
|
unknown |
-0.198 |
0.099 |
| 149 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
-0.197 |
0.092 |
| 150 |
264682_at |
AT1G65570
|
[Pectin lyase-like superfamily protein] |
-0.197 |
0.064 |
| 151 |
258064_at |
AT3G14680
|
CYP72A14, cytochrome P450, family 72, subfamily A, polypeptide 14 |
-0.192 |
0.123 |
| 152 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.191 |
0.098 |
| 153 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
-0.187 |
0.134 |
| 154 |
254215_at |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
-0.187 |
0.257 |
| 155 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.187 |
0.080 |
| 156 |
247005_at |
AT5G67520
|
adenosine-5'-phosphosulfate (APS) kinase 4 |
-0.186 |
-0.012 |
| 157 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
-0.185 |
0.037 |
| 158 |
254681_at |
AT4G18140
|
SSP4b, SCP1-like small phosphatase 4b |
-0.184 |
-0.007 |
| 159 |
247710_at |
AT5G59260
|
LecRK-II.1, L-type lectin receptor kinase II.1 |
-0.180 |
0.015 |
| 160 |
265995_at |
AT2G24140
|
unknown |
-0.178 |
0.064 |
| 161 |
253994_at |
AT4G26080
|
ABI1, ABA INSENSITIVE 1, AtABI1 |
-0.175 |
0.068 |
| 162 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
-0.175 |
0.060 |
| 163 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.174 |
0.058 |
| 164 |
253997_at |
AT4G26090
|
RPS2, RESISTANT TO P. SYRINGAE 2 |
-0.174 |
0.061 |
| 165 |
246195_at |
AT4G36410
|
UBC17, ubiquitin-conjugating enzyme 17 |
-0.172 |
0.197 |