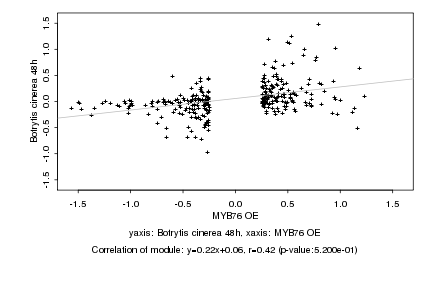
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
MYB76 OE |
Log2 signal ratio
Botrytis cinerea 48h |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.230 |
0.113 |
| 2 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.176 |
0.647 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
1.157 |
-0.507 |
| 4 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.130 |
-0.127 |
| 5 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
1.108 |
-0.193 |
| 6 |
257421_at |
AT1G12030
|
unknown |
0.997 |
0.032 |
| 7 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.967 |
-0.240 |
| 8 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.951 |
1.027 |
| 9 |
250589_at |
AT5G07700
|
AtMYB76, myb domain protein 76, MYB76, myb domain protein 76 |
0.947 |
0.050 |
| 10 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.945 |
0.082 |
| 11 |
250296_at |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
0.935 |
0.392 |
| 12 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.921 |
-0.211 |
| 13 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.845 |
0.195 |
| 14 |
250365_at |
AT5G11410
|
[Protein kinase superfamily protein] |
0.819 |
-0.055 |
| 15 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.818 |
0.339 |
| 16 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.799 |
0.350 |
| 17 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.783 |
1.487 |
| 18 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.772 |
0.857 |
| 19 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.762 |
0.798 |
| 20 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.725 |
-0.063 |
| 21 |
261794_at |
AT1G16060
|
ADAP, ARIA-interacting double AP2 domain protein, WRI3, WRINKLED 3 |
0.723 |
-0.090 |
| 22 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.720 |
0.140 |
| 23 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.718 |
0.042 |
| 24 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
0.705 |
-0.035 |
| 25 |
262118_at |
AT1G02850
|
BGLU11, beta glucosidase 11 |
0.704 |
0.432 |
| 26 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.692 |
0.331 |
| 27 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.669 |
-0.072 |
| 28 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.669 |
0.180 |
| 29 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.664 |
-0.009 |
| 30 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.651 |
1.013 |
| 31 |
267168_at |
AT2G37770
|
AKR4C9, Aldo-keto reductase family 4 member C9, ChlAKR, Chloroplastic aldo-keto reductase |
0.641 |
0.900 |
| 32 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.621 |
0.266 |
| 33 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.573 |
0.128 |
| 34 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
0.572 |
-0.182 |
| 35 |
264957_at |
AT1G77000
|
ATSKP2;2, ARABIDOPSIS HOMOLOG OF HOMOLOG OF HUMAN SKP2 2, SKP2B |
0.562 |
0.143 |
| 36 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
0.562 |
-0.143 |
| 37 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.553 |
0.190 |
| 38 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.552 |
0.169 |
| 39 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.549 |
-0.002 |
| 40 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.545 |
-0.011 |
| 41 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.544 |
0.729 |
| 42 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.539 |
0.008 |
| 43 |
252357_at |
AT3G48410
|
[alpha/beta-Hydrolases superfamily protein] |
0.539 |
0.032 |
| 44 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.531 |
1.247 |
| 45 |
254201_at |
AT4G24130
|
unknown |
0.523 |
0.068 |
| 46 |
258008_at |
AT3G19430
|
[late embryogenesis abundant protein-related / LEA protein-related] |
0.521 |
0.080 |
| 47 |
245254_at |
AT4G14680
|
APS3 |
0.520 |
0.137 |
| 48 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.511 |
1.112 |
| 49 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.501 |
0.216 |
| 50 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.495 |
1.143 |
| 51 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.488 |
-0.014 |
| 52 |
253913_at |
AT4G27370
|
ATVIIIB, MYOSIN VIII B, VIIIB |
0.481 |
-0.093 |
| 53 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.481 |
0.353 |
| 54 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.480 |
-0.025 |
| 55 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.477 |
-0.134 |
| 56 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
0.477 |
-0.009 |
| 57 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.467 |
0.139 |
| 58 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.466 |
-0.088 |
| 59 |
255471_at |
AT4G03050
|
AOP3 |
0.465 |
0.078 |
| 60 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.456 |
0.705 |
| 61 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
0.452 |
0.150 |
| 62 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
0.451 |
0.343 |
| 63 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.447 |
0.004 |
| 64 |
251037_at |
AT5G02100
|
ORP3A, OSBP(OXYSTEROL BINDING PROTEIN)-RELATED PROTEIN 3A, UNE18, UNFERTILIZED EMBRYO SAC 18 |
0.444 |
0.250 |
| 65 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.443 |
-0.215 |
| 66 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
0.438 |
0.384 |
| 67 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.433 |
0.429 |
| 68 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
0.430 |
0.103 |
| 69 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.422 |
0.239 |
| 70 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.405 |
-0.182 |
| 71 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
0.405 |
0.110 |
| 72 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.403 |
0.158 |
| 73 |
259911_at |
AT1G72680
|
ATCAD1, CINNAMYL ALCOHOL DEHYDROGENASE 1, CAD1, cinnamyl-alcohol dehydrogenase |
0.402 |
0.433 |
| 74 |
255306_at |
AT4G04740
|
ATCPK23, CPK23, calcium-dependent protein kinase 23 |
0.401 |
-0.123 |
| 75 |
264369_at |
AT1G70430
|
[Protein kinase superfamily protein] |
0.400 |
0.005 |
| 76 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.399 |
0.091 |
| 77 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.399 |
0.319 |
| 78 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
0.396 |
0.432 |
| 79 |
251515_at |
AT3G59270
|
[FBD-like domain family protein] |
0.391 |
0.008 |
| 80 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
0.390 |
0.344 |
| 81 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.387 |
0.495 |
| 82 |
246464_at |
AT5G16980
|
[Zinc-binding dehydrogenase family protein] |
0.385 |
0.534 |
| 83 |
255811_at |
AT4G10250
|
ATHSP22.0 |
0.384 |
0.061 |
| 84 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.381 |
0.769 |
| 85 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
0.376 |
-0.237 |
| 86 |
253689_at |
AT4G29770
|
unknown |
0.374 |
0.020 |
| 87 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
0.372 |
-0.185 |
| 88 |
246683_at |
AT5G33300
|
[chromosome-associated kinesin-related] |
0.371 |
0.079 |
| 89 |
263515_at |
AT2G21640
|
unknown |
0.367 |
0.635 |
| 90 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.358 |
0.282 |
| 91 |
246325_at |
AT1G16540
|
ABA3, ABA DEFICIENT 3, ACI2, ALTERED CHLOROPLAST IMPORT 2, ATABA3, AtLOS5, LOS5, LOW OSMOTIC STRESS 5, SIR3, SIRTINOL RESISTANT 3 |
0.354 |
0.486 |
| 92 |
255639_at |
AT4G00760
|
APRR8, pseudo-response regulator 8, PRR8, PSEUDO-RESPONSE REGULATOR 8 |
0.352 |
0.008 |
| 93 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.351 |
0.660 |
| 94 |
256021_at |
AT1G58270
|
ZW9 |
0.350 |
-0.032 |
| 95 |
246042_at |
AT5G19440
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.349 |
0.305 |
| 96 |
262380_at |
AT1G72810
|
Pyridoxal-5'-phosphate-dependent enzyme family protein |
0.348 |
-0.131 |
| 97 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
0.346 |
0.268 |
| 98 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.345 |
0.295 |
| 99 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.343 |
0.316 |
| 100 |
246418_at |
AT5G16960
|
[Zinc-binding dehydrogenase family protein] |
0.341 |
0.192 |
| 101 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.337 |
0.121 |
| 102 |
251147_at |
AT3G63480
|
[ATP binding microtubule motor family protein] |
0.337 |
0.105 |
| 103 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
0.331 |
0.196 |
| 104 |
258692_at |
AT3G08640
|
RER3, RETICULATA-RELATED 3 |
0.321 |
0.238 |
| 105 |
261460_at |
AT1G07880
|
ATMPK13 |
0.321 |
-0.052 |
| 106 |
250369_at |
AT5G11300
|
CYC2BAT, CYC3B, mitotic-like cyclin 3B from Arabidopsis, CYCA2;2, CYCLIN A2;2 |
0.317 |
-0.025 |
| 107 |
252274_at |
AT3G49680
|
ATBCAT-3, BCAT3, branched-chain aminotransferase 3 |
0.315 |
-0.101 |
| 108 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.314 |
0.392 |
| 109 |
249025_at |
AT5G44720
|
[Molybdenum cofactor sulfurase family protein] |
0.313 |
0.121 |
| 110 |
267112_at |
AT2G14750
|
AKN1, APS KINASE 1, APK, APS kinase, APK1, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 1, ATAKN1 |
0.309 |
0.081 |
| 111 |
263210_at |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.308 |
1.207 |
| 112 |
266915_at |
AT2G45870
|
[Bestrophin-like protein] |
0.304 |
0.049 |
| 113 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.299 |
-0.026 |
| 114 |
260385_at |
AT1G74090
|
ATSOT18, DESULFO-GLUCOSINOLATE SULFOTRANSFERASE 18, ATST5B, ARABIDOPSIS SULFOTRANSFERASE 5B, SOT18, desulfo-glucosinolate sulfotransferase 18 |
0.297 |
-0.026 |
| 115 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.297 |
0.066 |
| 116 |
259643_at |
AT1G68890
|
PHYLLO, PHYLLO |
0.296 |
-0.214 |
| 117 |
266627_at |
AT2G35340
|
MEE29, maternal effect embryo arrest 29 |
0.293 |
-0.178 |
| 118 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.291 |
0.254 |
| 119 |
246790_at |
AT5G27610
|
ALY1, ALWAYS EARLY 1, ATALY1, ARABIDOPSIS THALIANA ALWAYS EARLY 1 |
0.290 |
0.148 |
| 120 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.285 |
0.317 |
| 121 |
264855_at |
AT2G17265
|
DMR1, DOWNY MILDEW RESISTANT 1, HSK, homoserine kinase |
0.285 |
0.006 |
| 122 |
249638_at |
AT5G36880
|
ACS, acetyl-CoA synthetase |
0.283 |
0.196 |
| 123 |
261585_at |
AT1G01010
|
ANAC001, NAC domain containing protein 1, NAC001, NAC domain containing protein 1 |
0.279 |
0.102 |
| 124 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.279 |
0.177 |
| 125 |
255961_at |
AT1G22340
|
AtUGT85A7, UDP-glucosyl transferase 85A7, UGT85A7, UDP-glucosyl transferase 85A7 |
0.279 |
0.321 |
| 126 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
0.278 |
-0.009 |
| 127 |
254318_at |
AT4G22530
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.277 |
0.371 |
| 128 |
257716_at |
AT3G18300
|
unknown |
0.275 |
0.032 |
| 129 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.274 |
0.509 |
| 130 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.273 |
0.717 |
| 131 |
246043_at |
AT5G19380
|
CLT1, CRT (chloroquine-resistance transporter)-like transporter 1 |
0.272 |
0.062 |
| 132 |
259780_at |
AT1G29630
|
[5'-3' exonuclease family protein] |
0.271 |
0.096 |
| 133 |
257787_at |
AT3G27000
|
ARP2, actin related protein 2, ATARP2, ACTIN RELATED PROTEIN 2, WRM, WURM |
0.270 |
0.079 |
| 134 |
265276_at |
AT2G28400
|
unknown |
0.269 |
0.447 |
| 135 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
0.269 |
-0.039 |
| 136 |
250449_at |
AT5G10830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.269 |
0.027 |
| 137 |
263520_at |
AT2G42640
|
[Mitogen activated protein kinase kinase kinase-related] |
0.268 |
-0.103 |
| 138 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.267 |
0.056 |
| 139 |
261398_at |
AT1G79610
|
ATNHX6, ARABIDOPSIS NA+/H+ ANTIPORTER 6, NHX6, Na+/H+ antiporter 6 |
0.263 |
0.262 |
| 140 |
261642_at |
AT1G27680
|
APL2, ADPGLC-PPase large subunit |
0.262 |
-0.094 |
| 141 |
253343_at |
AT4G33540
|
[metallo-beta-lactamase family protein] |
0.262 |
0.456 |
| 142 |
259095_at |
AT3G05020
|
ACP, ACYL CARRIER PROTEIN, ACP1, acyl carrier protein 1 |
0.260 |
0.039 |
| 143 |
251262_at |
AT3G62080
|
[SNF7 family protein] |
0.260 |
-0.058 |
| 144 |
254742_at |
AT4G13430
|
ATLEUC1, IIL1, isopropyl malate isomerase large subunit 1 |
0.259 |
-0.012 |
| 145 |
251304_at |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
0.258 |
0.387 |
| 146 |
262607_at |
AT1G13990
|
unknown |
0.256 |
0.276 |
| 147 |
262752_at |
AT1G16330
|
CYCB3;1, cyclin b3;1 |
0.255 |
0.000 |
| 148 |
253884_at |
AT4G27670
|
HSP21, heat shock protein 21 |
0.255 |
0.148 |
| 149 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.254 |
0.016 |
| 150 |
258581_at |
AT3G04160
|
unknown |
0.253 |
-0.025 |
| 151 |
256510_at |
AT1G33360
|
[ATP-dependent Clp protease] |
0.252 |
0.306 |
| 152 |
252628_at |
AT3G44960
|
unknown |
0.251 |
0.071 |
| 153 |
263489_at |
AT2G31830
|
5PTase14, inositol-polyphosphate 5-phosphatase 14 |
0.249 |
-0.022 |
| 154 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-1.571 |
-0.118 |
| 155 |
245013_at |
ATCG00470
|
ATPE, ATP synthase epsilon chain |
-1.506 |
-0.005 |
| 156 |
244960_at |
ATCG01020
|
RPL32, ribosomal protein L32 |
-1.495 |
-0.019 |
| 157 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-1.478 |
-0.141 |
| 158 |
244969_at |
ATCG00650
|
RPS18, ribosomal protein S18 |
-1.379 |
-0.266 |
| 159 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-1.353 |
-0.116 |
| 160 |
244935_at |
ATCG01090
|
NDHI |
-1.277 |
-0.028 |
| 161 |
245021_at |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-1.245 |
0.011 |
| 162 |
244980_at |
ATCG00760
|
RPL36, ribosomal protein L36 |
-1.196 |
-0.030 |
| 163 |
244966_at |
ATCG00600
|
PETG |
-1.134 |
-0.075 |
| 164 |
244936_at |
ATCG01100
|
NDHA |
-1.110 |
-0.091 |
| 165 |
245014_at |
ATCG00480
|
ATPB, ATP synthase subunit beta, PB, ATP synthase subunit beta |
-1.061 |
0.004 |
| 166 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-1.052 |
-0.035 |
| 167 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-1.026 |
-0.130 |
| 168 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-1.024 |
-0.215 |
| 169 |
245001_at |
ATCG00220
|
PSBM, photosystem II reaction center protein M |
-1.018 |
0.022 |
| 170 |
244983_at |
ATCG00790
|
RPL16, ribosomal protein L16 |
-1.015 |
-0.081 |
| 171 |
244981_at |
ATCG00770
|
RPS8, ribosomal protein S8 |
-1.004 |
-0.066 |
| 172 |
245006_at |
ATCG00340
|
PSAB |
-0.998 |
0.010 |
| 173 |
244976_at |
ATCG00720
|
PETB, photosynthetic electron transfer B |
-0.996 |
-0.024 |
| 174 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
-0.985 |
-0.061 |
| 175 |
245004_at |
ATCG00300
|
YCF9 |
-0.861 |
-0.076 |
| 176 |
265892_at |
AT2G15020
|
unknown |
-0.838 |
-0.241 |
| 177 |
244979_at |
ATCG00750
|
RPS11, ribosomal protein S11 |
-0.810 |
-0.025 |
| 178 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
-0.800 |
-0.089 |
| 179 |
245022_at |
ATCG00560
|
PSBL, photosystem II reaction center protein L |
-0.798 |
0.001 |
| 180 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.751 |
-0.008 |
| 181 |
266516_at |
AT2G47880
|
[Glutaredoxin family protein] |
-0.747 |
-0.142 |
| 182 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.745 |
-0.418 |
| 183 |
267459_at |
AT2G33850
|
unknown |
-0.739 |
0.008 |
| 184 |
246947_at |
AT5G25120
|
CYP71B11, ytochrome p450, family 71, subfamily B, polypeptide 11 |
-0.707 |
-0.287 |
| 185 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.690 |
0.051 |
| 186 |
244967_at |
ATCG00630
|
PSAJ |
-0.687 |
-0.012 |
| 187 |
245012_at |
ATCG00440
|
NDHC |
-0.682 |
0.006 |
| 188 |
245047_at |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.676 |
-0.039 |
| 189 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.672 |
0.000 |
| 190 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.662 |
-0.513 |
| 191 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.658 |
-0.677 |
| 192 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.645 |
0.009 |
| 193 |
244978_at |
ATCG00740
|
RPOA, RNA polymerase subunit alpha |
-0.621 |
-0.008 |
| 194 |
245023_at |
ATCG00080
|
PSBI, photosystem II reaction center protein I |
-0.604 |
-0.034 |
| 195 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.601 |
0.492 |
| 196 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.594 |
-0.203 |
| 197 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.581 |
0.029 |
| 198 |
261221_at |
AT1G19960
|
unknown |
-0.573 |
-0.147 |
| 199 |
257334_at |
ATMG01370
|
ORF111D |
-0.558 |
0.048 |
| 200 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.550 |
-0.015 |
| 201 |
245776_at |
AT1G30260
|
unknown |
-0.541 |
-0.074 |
| 202 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.539 |
-0.216 |
| 203 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
-0.529 |
0.133 |
| 204 |
263480_at |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-0.528 |
-0.012 |
| 205 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.525 |
-0.102 |
| 206 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.514 |
-0.235 |
| 207 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.504 |
0.069 |
| 208 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.495 |
0.024 |
| 209 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.480 |
-0.161 |
| 210 |
264843_at |
AT1G03400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.467 |
-0.126 |
| 211 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.465 |
0.037 |
| 212 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.458 |
-0.683 |
| 213 |
249932_at |
AT5G22390
|
unknown |
-0.454 |
-0.489 |
| 214 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.453 |
-0.020 |
| 215 |
246860_at |
AT5G25840
|
unknown |
-0.447 |
-0.203 |
| 216 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.444 |
-0.132 |
| 217 |
247713_at |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.430 |
0.006 |
| 218 |
248007_at |
AT5G56260
|
[Ribonuclease E inhibitor RraA/Dimethylmenaquinone methyltransferase] |
-0.427 |
0.019 |
| 219 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.426 |
0.265 |
| 220 |
261407_at |
AT1G18810
|
[phytochrome kinase substrate-related] |
-0.423 |
-0.306 |
| 221 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.421 |
-0.567 |
| 222 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
-0.419 |
0.101 |
| 223 |
246146_at |
AT5G20050
|
[Protein kinase superfamily protein] |
-0.416 |
0.052 |
| 224 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.415 |
-0.142 |
| 225 |
267550_at |
AT2G32800
|
AP4.3A, LecRK-S.2, L-type lectin receptor kinase S.2 |
-0.413 |
0.050 |
| 226 |
254551_at |
AT4G19840
|
ATPP2-A1, phloem protein 2-A1, ATPP2A-1, phloem protein 2-A1, PP2-A1, phloem protein 2-A1 |
-0.411 |
-0.060 |
| 227 |
251916_at |
AT3G53960
|
[Major facilitator superfamily protein] |
-0.406 |
0.120 |
| 228 |
255645_at |
AT4G00880
|
SAUR31, SMALL AUXIN UPREGULATED RNA 31 |
-0.402 |
-0.201 |
| 229 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.397 |
-0.150 |
| 230 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.396 |
-0.076 |
| 231 |
260166_at |
AT1G79840
|
GL2, GLABRA 2 |
-0.394 |
-0.048 |
| 232 |
247592_at |
AT5G60780
|
ATNRT2.3, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.3, NRT2.3, nitrate transporter 2.3 |
-0.390 |
0.030 |
| 233 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.389 |
0.065 |
| 234 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.388 |
-0.227 |
| 235 |
250327_at |
AT5G12050
|
unknown |
-0.386 |
-0.306 |
| 236 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.383 |
-0.674 |
| 237 |
255957_at |
AT1G22160
|
unknown |
-0.378 |
-0.311 |
| 238 |
257333_at |
ATMG01360
|
COX1, cytochrome oxidase |
-0.374 |
0.122 |
| 239 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.372 |
0.356 |
| 240 |
260948_at |
AT1G06100
|
[Fatty acid desaturase family protein] |
-0.371 |
-0.067 |
| 241 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.361 |
-0.133 |
| 242 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
-0.359 |
0.063 |
| 243 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.359 |
-0.289 |
| 244 |
264310_at |
AT1G62030
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.359 |
-0.127 |
| 245 |
247693_at |
AT5G59730
|
ATEXO70H7, exocyst subunit exo70 family protein H7, EXO70H7, exocyst subunit exo70 family protein H7 |
-0.358 |
0.056 |
| 246 |
246687_at |
AT5G33370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.353 |
-0.011 |
| 247 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.341 |
0.248 |
| 248 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
-0.338 |
0.036 |
| 249 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.337 |
0.445 |
| 250 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.336 |
-0.329 |
| 251 |
252300_at |
AT3G49160
|
[pyruvate kinase family protein] |
-0.336 |
0.384 |
| 252 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-0.330 |
0.276 |
| 253 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.327 |
0.195 |
| 254 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-0.326 |
-0.717 |
| 255 |
256799_at |
AT3G18560
|
unknown |
-0.323 |
0.188 |
| 256 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.322 |
-0.265 |
| 257 |
251472_at |
AT3G59580
|
NLP9, NIN-like protein 9 |
-0.319 |
0.024 |
| 258 |
266808_at |
AT2G29995
|
unknown |
-0.315 |
0.056 |
| 259 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
-0.314 |
0.132 |
| 260 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
-0.305 |
-0.066 |
| 261 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.304 |
-0.488 |
| 262 |
263788_at |
AT2G24580
|
[FAD-dependent oxidoreductase family protein] |
-0.302 |
-0.282 |
| 263 |
255285_at |
AT4G04630
|
unknown |
-0.300 |
-0.128 |
| 264 |
247055_at |
AT5G66740
|
unknown |
-0.300 |
-0.074 |
| 265 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.298 |
-0.013 |
| 266 |
262598_at |
AT1G15260
|
unknown |
-0.296 |
-0.492 |
| 267 |
258225_at |
AT3G15630
|
unknown |
-0.294 |
-0.068 |
| 268 |
247980_at |
AT5G56860
|
GATA21, GATA TRANSCRIPTION FACTOR 21, GNC, GATA, nitrate-inducible, carbon metabolism-involved |
-0.294 |
-0.446 |
| 269 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.293 |
-0.432 |
| 270 |
264606_at |
AT1G04660
|
[glycine-rich protein] |
-0.291 |
0.057 |
| 271 |
248942_at |
AT5G45480
|
unknown |
-0.290 |
-0.011 |
| 272 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.286 |
0.019 |
| 273 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
-0.285 |
-0.016 |
| 274 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.281 |
0.108 |
| 275 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.281 |
-0.403 |
| 276 |
264463_at |
AT1G10150
|
[Carbohydrate-binding protein] |
-0.280 |
-0.012 |
| 277 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-0.279 |
-0.151 |
| 278 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.279 |
-0.399 |
| 279 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.278 |
0.006 |
| 280 |
263067_at |
AT2G17550
|
TRM26, TON1 Recruiting Motif 26 |
-0.278 |
-0.158 |
| 281 |
251733_at |
AT3G56240
|
CCH, copper chaperone |
-0.276 |
-0.049 |
| 282 |
263915_at |
AT2G36430
|
unknown |
-0.275 |
-0.384 |
| 283 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.271 |
-0.971 |
| 284 |
260058_at |
AT1G78100
|
AUF1, auxin up-regulated f-box protein 1 |
-0.270 |
-0.223 |
| 285 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
-0.270 |
-0.054 |
| 286 |
266824_at |
AT2G22800
|
HAT9 |
-0.269 |
0.015 |
| 287 |
262624_at |
AT1G06450
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.267 |
0.087 |
| 288 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
-0.265 |
0.090 |
| 289 |
257750_at |
AT3G18800
|
unknown |
-0.265 |
-0.209 |
| 290 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
-0.265 |
0.183 |
| 291 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.265 |
-0.542 |
| 292 |
251769_at |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
-0.264 |
0.454 |
| 293 |
265842_at |
AT2G35700
|
ATERF38, ERF FAMILY PROTEIN 38, ERF38, ERF family protein 38 |
-0.264 |
0.013 |
| 294 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.263 |
-0.397 |
| 295 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.262 |
0.063 |
| 296 |
259520_at |
AT1G12320
|
unknown |
-0.261 |
0.195 |
| 297 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.261 |
-0.075 |
| 298 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.261 |
-0.456 |
| 299 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
-0.261 |
0.439 |
| 300 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.260 |
-0.348 |
| 301 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.259 |
0.013 |
| 302 |
260302_at |
AT1G80310
|
MOT2, molybdate transporter 2 |
-0.258 |
0.050 |
| 303 |
264741_at |
AT1G62290
|
[Saposin-like aspartyl protease family protein] |
-0.258 |
-0.048 |
| 304 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.256 |
-0.103 |
| 305 |
247072_at |
AT5G66490
|
unknown |
-0.253 |
-0.155 |