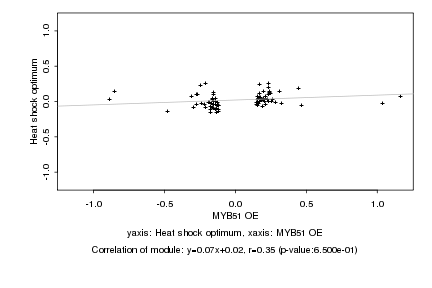
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
MYB51 OE |
Log2 signal ratio
Heat shock optimum |
| 1 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
1.161 |
0.073 |
| 2 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
1.036 |
-0.014 |
| 3 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
0.461 |
-0.047 |
| 4 |
252149_at |
AT3G51290
|
APSR1, Altered Phosphate Starvation Response 1 |
0.440 |
0.186 |
| 5 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.319 |
-0.024 |
| 6 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.308 |
0.154 |
| 7 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.279 |
-0.013 |
| 8 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
0.258 |
0.038 |
| 9 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
0.254 |
0.006 |
| 10 |
253162_at |
AT4G35630
|
PSAT1, phosphoserine aminotransferase 1 |
0.242 |
0.114 |
| 11 |
267144_at |
AT2G38110
|
ATGPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6, GPAT6, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 6 |
0.240 |
0.125 |
| 12 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.237 |
0.147 |
| 13 |
252093_at |
AT3G51500
|
unknown |
0.236 |
0.130 |
| 14 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
0.232 |
0.256 |
| 15 |
266476_at |
AT2G31090
|
unknown |
0.231 |
0.001 |
| 16 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.230 |
0.203 |
| 17 |
256064_at |
AT1G07020
|
unknown |
0.229 |
0.112 |
| 18 |
262485_at |
AT1G21730
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.220 |
0.042 |
| 19 |
257761_at |
AT3G23090
|
WDL3 |
0.210 |
0.054 |
| 20 |
254492_at |
AT4G20260
|
ATPCAP1, ARABIDOPSIS THALIANA PLASMA-MEMBRANE ASSOCIATED CATION-BINDING PROTEIN 1, MDP25, microtubule-destabilizing protein 25, PCAP1, plasma-membrane associated cation-binding protein 1 |
0.208 |
0.081 |
| 21 |
260913_at |
AT1G02500
|
AtSAM1, MAT1, METK1, SAM-1, S-ADENOSYLMETHIONINE SYNTHETASE-1, SAM1, S-adenosylmethionine synthetase 1 |
0.205 |
-0.029 |
| 22 |
253021_at |
AT4G38050
|
[Xanthine/uracil permease family protein] |
0.203 |
0.002 |
| 23 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.196 |
0.048 |
| 24 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.195 |
0.146 |
| 25 |
261510_at |
AT1G71760
|
unknown |
0.193 |
0.036 |
| 26 |
267431_at |
AT2G34870
|
MEE26, maternal effect embryo arrest 26 |
0.185 |
-0.064 |
| 27 |
259306_at |
AT3G05290
|
AtPNC1, PNC1, peroxisomal adenine nucleotide carrier 1 |
0.180 |
0.028 |
| 28 |
254845_at |
AT4G11820
|
FKP1, FLAKY POLLEN 1, HMGS, HYDROXYMETHYLGLUTARYL-COA SYNTHASE, MVA1 |
0.178 |
0.037 |
| 29 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
0.174 |
0.068 |
| 30 |
263609_at |
AT2G16250
|
[Leucine-rich repeat protein kinase family protein] |
0.171 |
0.054 |
| 31 |
258880_at |
AT3G06420
|
ATG8H, autophagy 8h |
0.169 |
0.065 |
| 32 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.168 |
0.251 |
| 33 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.167 |
0.126 |
| 34 |
247831_at |
AT5G58540
|
[Protein kinase superfamily protein] |
0.163 |
0.007 |
| 35 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.161 |
0.048 |
| 36 |
253661_at |
AT4G30130
|
unknown |
0.158 |
0.044 |
| 37 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
0.157 |
-0.021 |
| 38 |
263909_at |
AT2G36490
|
AtROS1, DML1, demeter-like 1, ROS1, REPRESSOR OF SILENCING1 |
0.155 |
-0.012 |
| 39 |
258132_at |
AT3G24550
|
ATPERK1, proline-rich extensin-like receptor kinase 1, PERK1, proline-rich extensin-like receptor kinase 1 |
0.154 |
0.042 |
| 40 |
261039_at |
AT1G17455
|
ELF4-L4, ELF4-like 4 |
0.153 |
0.075 |
| 41 |
261597_at |
AT1G49780
|
PUB26, plant U-box 26 |
0.151 |
-0.049 |
| 42 |
262277_at |
AT1G68650
|
[Uncharacterized protein family (UPF0016)] |
0.148 |
-0.037 |
| 43 |
262602_at |
AT1G15270
|
[Translation machinery associated TMA7] |
0.145 |
-0.004 |
| 44 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.893 |
0.040 |
| 45 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.855 |
0.142 |
| 46 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.484 |
-0.136 |
| 47 |
250935_at |
AT5G03240
|
UBQ3, polyubiquitin 3 |
-0.315 |
0.073 |
| 48 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
-0.297 |
-0.077 |
| 49 |
263352_at |
AT2G22080
|
unknown |
-0.280 |
-0.041 |
| 50 |
265025_at |
AT1G24575
|
unknown |
-0.275 |
0.105 |
| 51 |
263478_at |
AT2G31880
|
EVR, EVERSHED, SOBIR1, SUPPRESSOR OF BIR1 1 |
-0.270 |
0.106 |
| 52 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.251 |
0.237 |
| 53 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.240 |
-0.018 |
| 54 |
249529_at |
AT5G38730
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.225 |
-0.040 |
| 55 |
255842_at |
AT2G33530
|
scpl46, serine carboxypeptidase-like 46 |
-0.216 |
-0.084 |
| 56 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.215 |
0.268 |
| 57 |
263605_at |
AT2G16485
|
NERD, Needed for RDR2-independent DNA methylation |
-0.197 |
-0.005 |
| 58 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.183 |
-0.009 |
| 59 |
247243_at |
AT5G64700
|
UMAMIT21, Usually multiple acids move in and out Transporters 21 |
-0.180 |
-0.057 |
| 60 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.178 |
-0.106 |
| 61 |
253517_at |
AT4G31390
|
ABC1K1, ABC1-like kinase 1, ACDO1, ABC1-like kinase related to chlorophyll degradation and oxidative stress 1, AtACDO1, PGR6, PROTON GRADIENT REGULATION 6 |
-0.178 |
-0.149 |
| 62 |
249482_at |
AT5G38980
|
unknown |
-0.172 |
-0.028 |
| 63 |
262131_at |
AT1G02900
|
ATRALF1, RAPID ALKALINIZATION FACTOR 1, RALF1, rapid alkalinization factor 1, RALFL1, RALF-LIKE 1 |
-0.168 |
0.030 |
| 64 |
263553_at |
AT2G16430
|
ATPAP10, PAP10, purple acid phosphatase 10 |
-0.167 |
-0.084 |
| 65 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.165 |
0.045 |
| 66 |
261463_at |
AT1G07740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.160 |
-0.032 |
| 67 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.158 |
0.140 |
| 68 |
262560_at |
AT1G34280
|
unknown |
-0.158 |
-0.097 |
| 69 |
262529_at |
AT1G17250
|
AtRLP3, receptor like protein 3, RFO2, RESISTANCE TO FUSARIUM OXYSPORUM 2, RLP3, receptor like protein 3 |
-0.157 |
0.013 |
| 70 |
262750_at |
AT1G28710
|
[Nucleotide-diphospho-sugar transferase family protein] |
-0.157 |
-0.089 |
| 71 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
-0.155 |
0.107 |
| 72 |
250990_at |
AT5G02290
|
NAK |
-0.147 |
0.052 |
| 73 |
263007_at |
AT1G54260
|
[winged-helix DNA-binding transcription factor family protein] |
-0.144 |
-0.104 |
| 74 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
-0.143 |
-0.039 |
| 75 |
251002_at |
AT5G02680
|
[LOCATED IN: endomembrane system] |
-0.141 |
0.008 |
| 76 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.139 |
-0.147 |
| 77 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.138 |
-0.085 |
| 78 |
259475_at |
AT1G19140
|
unknown |
-0.135 |
-0.088 |
| 79 |
262365_at |
AT1G72870
|
[Disease resistance protein (TIR-NBS class)] |
-0.133 |
-0.045 |
| 80 |
265472_at |
AT2G15580
|
[RING/U-box superfamily protein] |
-0.132 |
-0.031 |
| 81 |
260748_at |
AT1G49210
|
[RING/U-box superfamily protein] |
-0.127 |
-0.004 |
| 82 |
249386_at |
AT5G40060
|
[Disease resistance protein (NBS-LRR class) family] |
-0.126 |
-0.102 |
| 83 |
256519_at |
AT1G66110
|
unknown |
-0.125 |
-0.043 |
| 84 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
-0.124 |
-0.136 |
| 85 |
260916_at |
AT1G02475
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.123 |
-0.048 |