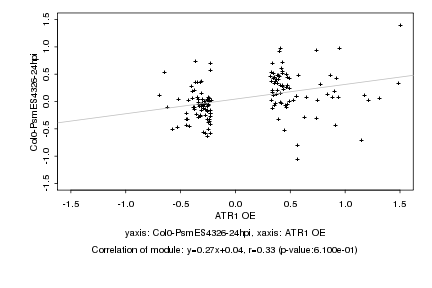
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ATR1 OE |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
1.501 |
1.398 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.484 |
0.334 |
| 3 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
1.306 |
0.071 |
| 4 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.206 |
0.035 |
| 5 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
1.176 |
0.125 |
| 6 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
1.148 |
-0.707 |
| 7 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.946 |
0.984 |
| 8 |
254201_at |
AT4G24130
|
unknown |
0.934 |
0.077 |
| 9 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.916 |
0.436 |
| 10 |
262646_at |
AT1G62800
|
ASP4, aspartate aminotransferase 4 |
0.909 |
-0.428 |
| 11 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.895 |
0.187 |
| 12 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.876 |
0.085 |
| 13 |
262607_at |
AT1G13990
|
unknown |
0.860 |
0.492 |
| 14 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.835 |
0.129 |
| 15 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.774 |
0.317 |
| 16 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.739 |
0.034 |
| 17 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.732 |
-0.308 |
| 18 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.732 |
0.946 |
| 19 |
261181_at |
AT1G34580
|
[Major facilitator superfamily protein] |
0.646 |
0.091 |
| 20 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.628 |
-0.291 |
| 21 |
259596_at |
AT1G28130
|
GH3.17 |
0.572 |
0.488 |
| 22 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.560 |
-0.799 |
| 23 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.557 |
-1.046 |
| 24 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.555 |
0.104 |
| 25 |
266525_at |
AT2G16970
|
MEE15, maternal effect embryo arrest 15 |
0.553 |
0.092 |
| 26 |
266915_at |
AT2G45870
|
[Bestrophin-like protein] |
0.525 |
0.019 |
| 27 |
267464_at |
AT2G19150
|
[Pectin lyase-like superfamily protein] |
0.492 |
0.017 |
| 28 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.489 |
0.436 |
| 29 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.484 |
0.248 |
| 30 |
246789_at |
AT5G27600
|
ATLACS7, LACS7, long-chain acyl-CoA synthetase 7 |
0.471 |
0.443 |
| 31 |
266167_at |
AT2G38860
|
DJ-1e, DJ1E, YLS5 |
0.466 |
0.305 |
| 32 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.466 |
-0.040 |
| 33 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.465 |
0.496 |
| 34 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
0.463 |
0.259 |
| 35 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.460 |
-0.096 |
| 36 |
263900_at |
AT2G36290
|
[alpha/beta-Hydrolases superfamily protein] |
0.455 |
-0.060 |
| 37 |
258038_at |
AT3G21260
|
GLTP3, GLYCOLIPID TRANSFER PROTEIN 3 |
0.447 |
-0.517 |
| 38 |
263098_at |
AT2G16005
|
[MD-2-related lipid recognition domain-containing protein] |
0.431 |
0.229 |
| 39 |
249910_at |
AT5G22630
|
ADT5, arogenate dehydratase 5 |
0.428 |
0.556 |
| 40 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.427 |
0.725 |
| 41 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
0.425 |
0.520 |
| 42 |
249372_at |
AT5G40760
|
G6PD6, glucose-6-phosphate dehydrogenase 6 |
0.424 |
0.288 |
| 43 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
0.423 |
0.299 |
| 44 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.418 |
0.610 |
| 45 |
254015_at |
AT4G26140
|
BGAL12, beta-galactosidase 12 |
0.418 |
-0.029 |
| 46 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.409 |
0.981 |
| 47 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.408 |
-0.013 |
| 48 |
263807_at |
AT2G04400
|
[Aldolase-type TIM barrel family protein] |
0.405 |
0.311 |
| 49 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.402 |
0.159 |
| 50 |
264580_at |
AT1G05340
|
unknown |
0.400 |
0.922 |
| 51 |
253332_at |
AT4G33420
|
[Peroxidase superfamily protein] |
0.393 |
0.475 |
| 52 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
0.392 |
0.483 |
| 53 |
255285_at |
AT4G04630
|
unknown |
0.392 |
-0.324 |
| 54 |
248004_at |
AT5G56230
|
PRA1.G2, prenylated RAB acceptor 1.G2 |
0.384 |
0.404 |
| 55 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.383 |
0.335 |
| 56 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.382 |
0.208 |
| 57 |
253343_at |
AT4G33540
|
[metallo-beta-lactamase family protein] |
0.382 |
0.483 |
| 58 |
259708_at |
AT1G77420
|
[alpha/beta-Hydrolases superfamily protein] |
0.373 |
0.409 |
| 59 |
262049_at |
AT1G80180
|
unknown |
0.367 |
0.141 |
| 60 |
252040_at |
AT3G52060
|
AtGnTL, GnTL, beta-1,6-N-acetylglucosaminyl transferase-like |
0.359 |
-0.025 |
| 61 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.358 |
0.368 |
| 62 |
264447_at |
AT1G27300
|
unknown |
0.356 |
0.456 |
| 63 |
258300_at |
AT3G23340
|
ckl10, casein kinase I-like 10 |
0.356 |
-0.061 |
| 64 |
261269_at |
AT1G26690
|
[emp24/gp25L/p24 family/GOLD family protein] |
0.352 |
0.333 |
| 65 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
0.344 |
0.438 |
| 66 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
0.344 |
0.111 |
| 67 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
0.339 |
0.521 |
| 68 |
258815_at |
AT3G04000
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.337 |
0.712 |
| 69 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
0.336 |
-0.119 |
| 70 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.331 |
0.216 |
| 71 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
0.329 |
0.180 |
| 72 |
262844_at |
AT1G14890
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.328 |
0.033 |
| 73 |
258021_at |
AT3G19380
|
PUB25, plant U-box 25 |
0.323 |
0.382 |
| 74 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.323 |
0.541 |
| 75 |
258275_at |
AT3G15760
|
unknown |
0.317 |
0.471 |
| 76 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
-0.698 |
0.118 |
| 77 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
-0.651 |
0.539 |
| 78 |
261684_at |
AT1G47400
|
unknown |
-0.622 |
-0.097 |
| 79 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.581 |
-0.494 |
| 80 |
258034_at |
AT3G21300
|
[RNA methyltransferase family protein] |
-0.534 |
-0.460 |
| 81 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.526 |
0.039 |
| 82 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.455 |
-0.432 |
| 83 |
251476_at |
AT3G59670
|
unknown |
-0.455 |
-0.317 |
| 84 |
248891_at |
AT5G46280
|
MCM3, MINICHROMOSOME MAINTENANCE 3 |
-0.454 |
-0.215 |
| 85 |
264188_at |
AT1G54690
|
G-H2AX, GAMMA H2AX, GAMMA-H2AX, gamma histone variant H2AX, H2AXB, HTA3, histone H2A 3 |
-0.438 |
-0.324 |
| 86 |
253788_at |
AT4G28680
|
AtTYDC, TYDC, L-tyrosine decarboxylase, TYRDC, L-tyrosine decarboxylase, TYRDC1, L-TYROSINE DECARBOXYLASE 1 |
-0.436 |
0.028 |
| 87 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
-0.419 |
-0.442 |
| 88 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.408 |
0.285 |
| 89 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
-0.400 |
0.057 |
| 90 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
-0.394 |
0.191 |
| 91 |
254452_at |
AT4G21100
|
DDB1B, damaged DNA binding protein 1B |
-0.386 |
-0.102 |
| 92 |
263352_at |
AT2G22080
|
unknown |
-0.381 |
-0.106 |
| 93 |
246303_at |
AT3G51870
|
[Mitochondrial substrate carrier family protein] |
-0.381 |
-0.132 |
| 94 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.381 |
0.218 |
| 95 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.373 |
0.359 |
| 96 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.372 |
0.750 |
| 97 |
256144_at |
AT1G48630
|
RACK1B, receptor for activated C kinase 1B, RACK1B_AT, receptor for activated C kinase 1B |
-0.357 |
-0.222 |
| 98 |
246920_at |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-0.353 |
0.076 |
| 99 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
-0.350 |
0.366 |
| 100 |
264262_at |
AT1G09200
|
H3.1, histone 3.1 |
-0.344 |
0.048 |
| 101 |
264061_at |
AT2G27970
|
CKS2, CDK-subunit 2 |
-0.341 |
-0.049 |
| 102 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.339 |
-0.014 |
| 103 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.337 |
-0.276 |
| 104 |
265097_at |
AT1G04020
|
ATBARD1, BARD1, breast cancer associated RING 1, ROW1 |
-0.329 |
-0.084 |
| 105 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.326 |
-0.264 |
| 106 |
247168_at |
AT5G65860
|
[ankyrin repeat family protein] |
-0.323 |
-0.244 |
| 107 |
266166_at |
AT2G28080
|
[UDP-Glycosyltransferase superfamily protein] |
-0.322 |
0.361 |
| 108 |
247192_at |
AT5G65360
|
H3.1, histone 3.1 |
-0.318 |
-0.089 |
| 109 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
-0.317 |
0.371 |
| 110 |
253535_at |
AT4G31550
|
ATWRKY11, WRKY11, WRKY DNA-binding protein 11 |
-0.315 |
0.153 |
| 111 |
262819_at |
AT1G11600
|
CYP77B1, cytochrome P450, family 77, subfamily B, polypeptide 1 |
-0.313 |
-0.050 |
| 112 |
260683_at |
AT1G17560
|
HLL, HUELLENLOS |
-0.310 |
-0.164 |
| 113 |
258089_at |
AT3G14740
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-0.308 |
-0.032 |
| 114 |
246315_at |
AT3G56870
|
unknown |
-0.306 |
0.048 |
| 115 |
247651_at |
AT5G59870
|
HTA6, histone H2A 6 |
-0.302 |
-0.064 |
| 116 |
263047_at |
AT2G17630
|
PSAT2, phosphoserine aminotransferase 2 |
-0.296 |
-0.112 |
| 117 |
257735_at |
AT3G27400
|
[Pectin lyase-like superfamily protein] |
-0.296 |
-0.549 |
| 118 |
261330_at |
AT1G44900
|
ATMCM2, MCM2, MINICHROMOSOME MAINTENANCE 2 |
-0.294 |
-0.090 |
| 119 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-0.291 |
0.008 |
| 120 |
246088_at |
AT5G20600
|
unknown |
-0.290 |
-0.038 |
| 121 |
251442_at |
AT3G59980
|
[Nucleic acid-binding, OB-fold-like protein] |
-0.282 |
-0.137 |
| 122 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
-0.279 |
-0.574 |
| 123 |
249276_at |
AT5G41880
|
POLA3, POLA4 |
-0.279 |
-0.029 |
| 124 |
253996_at |
AT4G26110
|
ATNAP1;1, ARABIDOPSIS THALIANA NUCLEOSOME ASSEMLY PROTEIN 1;1, NAP1;1, nucleosome assembly protein1;1 |
-0.276 |
-0.251 |
| 125 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
-0.263 |
-0.311 |
| 126 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
-0.261 |
0.047 |
| 127 |
258480_at |
AT3G02640
|
unknown |
-0.261 |
-0.176 |
| 128 |
258554_at |
AT3G06980
|
[DEA(D/H)-box RNA helicase family protein] |
-0.258 |
-0.634 |
| 129 |
252361_at |
AT3G48490
|
unknown |
-0.258 |
0.033 |
| 130 |
252661_at |
AT3G44450
|
unknown |
-0.258 |
-0.080 |
| 131 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
-0.256 |
-0.151 |
| 132 |
253490_at |
AT4G31790
|
[Tetrapyrrole (Corrin/Porphyrin) Methylases] |
-0.254 |
-0.039 |
| 133 |
252915_at |
AT4G38810
|
[Calcium-binding EF-hand family protein] |
-0.253 |
0.083 |
| 134 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
-0.252 |
-0.378 |
| 135 |
247482_at |
AT5G62410
|
ATCAP-E1, ATSMC4, SMC2, structural maintenance of chromosomes 2, TTN3, TITAN 3 |
-0.251 |
-0.070 |
| 136 |
258534_at |
AT3G06730
|
TRX P, thioredoxin putative plastidic, TRX z, Thioredoxin z |
-0.247 |
-0.511 |
| 137 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.244 |
0.003 |
| 138 |
246746_at |
AT5G27820
|
[Ribosomal L18p/L5e family protein] |
-0.242 |
-0.316 |
| 139 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.240 |
-0.353 |
| 140 |
256666_at |
AT3G20670
|
HTA13, histone H2A 13 |
-0.237 |
0.058 |
| 141 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.236 |
-0.264 |
| 142 |
266079_at |
AT2G37860
|
LCD1, LOWER CELL DENSITY 1 |
-0.235 |
-0.582 |
| 143 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.233 |
0.701 |
| 144 |
252147_at |
AT3G51270
|
[protein serine/threonine kinases] |
-0.233 |
-0.408 |
| 145 |
245404_at |
AT4G17610
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
-0.232 |
-0.151 |
| 146 |
263939_at |
AT2G36070
|
ATTIM44-2, translocase inner membrane subunit 44-2, TIM44-2, translocase inner membrane subunit 44-2 |
-0.232 |
-0.216 |
| 147 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
-0.231 |
0.575 |
| 148 |
245296_at |
AT4G16370
|
oligopeptide transporter |
-0.229 |
-0.218 |
| 149 |
249946_at |
AT5G19170
|
unknown |
-0.226 |
0.022 |