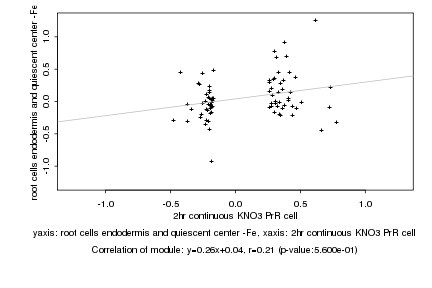
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr continuous KNO3 PrR cell |
Log2 signal ratio
root cells endodermis and quiescent center -Fe |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.773 |
-0.311 |
| 2 |
264014_at |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
0.726 |
0.222 |
| 3 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.717 |
-0.092 |
| 4 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.659 |
-0.441 |
| 5 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.615 |
1.268 |
| 6 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.504 |
-0.013 |
| 7 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.463 |
-0.095 |
| 8 |
265618_at |
AT2G25460
|
unknown |
0.456 |
0.385 |
| 9 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.438 |
-0.075 |
| 10 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.432 |
-0.214 |
| 11 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.421 |
0.149 |
| 12 |
259015_at |
AT3G07350
|
unknown |
0.414 |
0.459 |
| 13 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.404 |
0.023 |
| 14 |
248400_at |
AT5G52020
|
[Integrase-type DNA-binding superfamily protein] |
0.401 |
0.048 |
| 15 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.392 |
0.704 |
| 16 |
261842_at |
AT1G16160
|
WAKL5, wall associated kinase-like 5 |
0.371 |
-0.059 |
| 17 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.371 |
0.927 |
| 18 |
245779_at |
AT1G73510
|
unknown |
0.367 |
0.328 |
| 19 |
246054_at |
AT5G08360
|
unknown |
0.358 |
0.201 |
| 20 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.355 |
-0.103 |
| 21 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.342 |
-0.213 |
| 22 |
267263_at |
AT2G23110
|
[Late embryogenesis abundant protein, group 6] |
0.341 |
0.285 |
| 23 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.338 |
-0.193 |
| 24 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.332 |
-0.003 |
| 25 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.331 |
0.147 |
| 26 |
246279_at |
AT4G36740
|
ATHB40, homeobox protein 40, HB-5, HB40, homeobox protein 40 |
0.331 |
0.463 |
| 27 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
0.318 |
-0.071 |
| 28 |
265184_at |
AT1G23710
|
unknown |
0.313 |
0.690 |
| 29 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.307 |
-0.016 |
| 30 |
250073_at |
AT5G17170
|
ENH1, enhancer of sos3-1 |
0.305 |
0.012 |
| 31 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.300 |
-0.160 |
| 32 |
257840_at |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
0.296 |
0.365 |
| 33 |
247492_at |
AT5G61890
|
[Integrase-type DNA-binding superfamily protein] |
0.294 |
0.774 |
| 34 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.287 |
0.349 |
| 35 |
262284_at |
AT1G68670
|
[myb-like transcription factor family protein] |
0.278 |
0.094 |
| 36 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.273 |
-0.064 |
| 37 |
245593_at |
AT4G14550
|
IAA14, indole-3-acetic acid inducible 14, SLR, SOLITARY ROOT |
0.272 |
-0.026 |
| 38 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
0.270 |
0.211 |
| 39 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.259 |
-0.078 |
| 40 |
260522_x_at |
AT2G41730
|
unknown |
0.257 |
0.326 |
| 41 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
0.257 |
0.165 |
| 42 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
0.256 |
0.297 |
| 43 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.478 |
-0.284 |
| 44 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.425 |
0.456 |
| 45 |
249812_at |
AT5G23830
|
[MD-2-related lipid recognition domain-containing protein] |
-0.370 |
-0.043 |
| 46 |
254723_at |
AT4G13510
|
AMT1;1, ammonium transporter 1;1, ATAMT1, ARABIDOPSIS THALIANA AMMONIUM TRANSPORT 1, ATAMT1;1 |
-0.370 |
-0.299 |
| 47 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
-0.340 |
-0.114 |
| 48 |
252202_at |
AT3G50300
|
[HXXXD-type acyl-transferase family protein] |
-0.291 |
0.281 |
| 49 |
250104_at |
AT5G16610
|
unknown |
-0.279 |
0.264 |
| 50 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
-0.275 |
-0.241 |
| 51 |
245310_at |
AT4G13950
|
RALFL31, ralf-like 31 |
-0.264 |
-0.198 |
| 52 |
267148_at |
AT2G23470
|
RUS4, ROOT UV-B SENSITIVE 4 |
-0.259 |
-0.019 |
| 53 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.256 |
0.438 |
| 54 |
253112_at |
AT4G35970
|
APX5, ascorbate peroxidase 5 |
-0.236 |
-0.347 |
| 55 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
-0.231 |
0.015 |
| 56 |
254442_at |
AT4G21060
|
AtGALT2, GALT2, AGP galactosyltransferase 2 |
-0.229 |
0.114 |
| 57 |
259207_at |
AT3G09050
|
unknown |
-0.226 |
-0.122 |
| 58 |
245990_at |
AT5G20640
|
unknown |
-0.226 |
-0.292 |
| 59 |
265253_at |
AT2G02020
|
AtNPF8.4, AtPTR4, NPF8.4, NRT1/ PTR family 8.4, PTR4, peptide transporter 4 |
-0.221 |
-0.124 |
| 60 |
252621_at |
AT3G45270
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 5.7e-58 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
-0.215 |
-0.031 |
| 61 |
259462_at |
AT1G18940
|
[Nodulin-like / Major Facilitator Superfamily protein] |
-0.213 |
-0.294 |
| 62 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.210 |
0.076 |
| 63 |
251716_at |
AT3G55870
|
[ADC synthase superfamily protein] |
-0.207 |
0.152 |
| 64 |
261841_at |
AT1G15920
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.207 |
0.172 |
| 65 |
257805_at |
AT3G18830
|
ATPLT5, POLYOL TRANSPORTER 5, ATPMT5, ARABIDOPSIS THALIANA POLYOL/MONOSACCHARIDE TRANSPORTER 5, PMT5, polyol/monosaccharide transporter 5 |
-0.206 |
0.052 |
| 66 |
263970_at |
AT2G42850
|
CYP718, cytochrome P450, family 718 |
-0.204 |
-0.418 |
| 67 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.200 |
0.235 |
| 68 |
261085_at |
AT1G17480
|
IQD7, IQ-domain 7 |
-0.198 |
-0.102 |
| 69 |
265433_at |
AT2G20950
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
-0.198 |
-0.053 |
| 70 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.197 |
-0.183 |
| 71 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.194 |
-0.031 |
| 72 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
-0.192 |
-0.920 |
| 73 |
251820_at |
AT3G55040
|
GSTL2, glutathione transferase lambda 2 |
-0.192 |
-0.066 |
| 74 |
252901_at |
AT4G39550
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.190 |
-0.003 |
| 75 |
258470_at |
AT3G06035
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.188 |
-0.081 |
| 76 |
266068_at |
AT2G18640
|
GGPS4, geranylgeranyl pyrophosphate synthase 4 |
-0.186 |
-0.158 |
| 77 |
263719_at |
AT2G13600
|
SLO2, SLOW GROWTH 2 |
-0.186 |
0.046 |
| 78 |
251380_at |
AT3G60700
|
unknown |
-0.186 |
-0.003 |
| 79 |
254974_at |
AT4G10490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.184 |
0.049 |
| 80 |
251805_at |
AT3G55530
|
AtSDIR1, SDIR1, SALT- AND DROUGHT-INDUCED RING FINGER1 |
-0.183 |
-0.075 |
| 81 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.177 |
0.025 |
| 82 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.176 |
0.056 |
| 83 |
247020_at |
AT5G67020
|
unknown |
-0.175 |
0.484 |