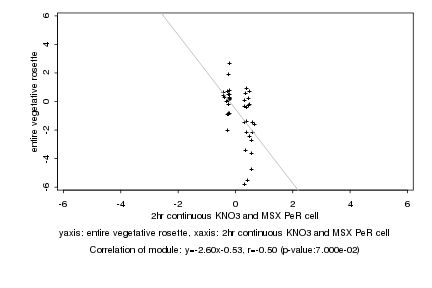
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr continuous KNO3 and MSX PeR cell |
Log2 signal ratio
entire vegetative rosette |
| 1 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.641 |
-1.543 |
| 2 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.590 |
-1.415 |
| 3 |
246142_at |
AT5G19970
|
unknown |
0.561 |
-2.144 |
| 4 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.558 |
-2.725 |
| 5 |
259813_at |
AT1G49860
|
ATGSTF14, glutathione S-transferase (class phi) 14, GSTF14, glutathione S-transferase (class phi) 14 |
0.558 |
-4.731 |
| 6 |
250472_at |
AT5G10210
|
unknown |
0.531 |
-3.586 |
| 7 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
0.485 |
0.747 |
| 8 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.474 |
-2.401 |
| 9 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.463 |
-0.195 |
| 10 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.450 |
0.273 |
| 11 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.421 |
-0.254 |
| 12 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.412 |
-5.524 |
| 13 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.379 |
-1.396 |
| 14 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.374 |
-2.170 |
| 15 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.363 |
0.929 |
| 16 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.359 |
-0.357 |
| 17 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
0.340 |
-3.373 |
| 18 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.340 |
0.632 |
| 19 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.303 |
-5.746 |
| 20 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.301 |
-0.297 |
| 21 |
251396_at |
AT3G60750
|
AtTKL1, TKL1, transketolase 1 |
0.300 |
0.122 |
| 22 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.299 |
-1.444 |
| 23 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.442 |
0.673 |
| 24 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
-0.429 |
0.485 |
| 25 |
260968_at |
AT1G12250
|
[Pentapeptide repeat-containing protein] |
-0.407 |
0.308 |
| 26 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.313 |
0.045 |
| 27 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.305 |
-0.845 |
| 28 |
261133_at |
AT1G19715
|
[Mannose-binding lectin superfamily protein] |
-0.296 |
0.765 |
| 29 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.279 |
-2.015 |
| 30 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.277 |
-0.150 |
| 31 |
257814_at |
AT3G25110
|
AtFaTA, fatA acyl-ACP thioesterase, FaTA, fatA acyl-ACP thioesterase |
-0.268 |
-0.806 |
| 32 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.258 |
-0.854 |
| 33 |
254193_at |
AT4G23870
|
unknown |
-0.253 |
0.495 |
| 34 |
267077_at |
AT2G40970
|
MYBC1 |
-0.251 |
0.141 |
| 35 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.243 |
1.962 |
| 36 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
-0.241 |
2.704 |
| 37 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.239 |
0.780 |
| 38 |
254209_at |
AT4G23490
|
unknown |
-0.233 |
0.219 |
| 39 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
-0.233 |
-0.786 |
| 40 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
-0.227 |
0.184 |
| 41 |
260540_at |
AT2G43500
|
NLP8, NIN-like protein 8 |
-0.225 |
0.284 |
| 42 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
-0.224 |
0.539 |
| 43 |
254412_at |
AT4G21430
|
B160 |
-0.220 |
0.219 |