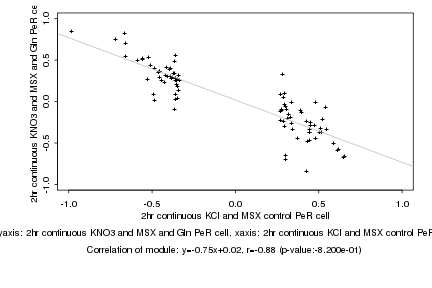
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr continuous KCl and MSX control PeR cell |
Log2 signal ratio
2hr continuous KNO3 and MSX and Gln PeR cell |
| 1 |
263658_at |
AT1G04490
|
unknown |
0.652 |
-0.653 |
| 2 |
251019_at |
AT5G02420
|
unknown |
0.643 |
-0.671 |
| 3 |
263575_at |
AT2G17070
|
unknown |
0.618 |
-0.567 |
| 4 |
252218_at |
AT3G50150
|
unknown |
0.608 |
-0.589 |
| 5 |
259027_at |
AT3G09280
|
unknown |
0.583 |
-0.502 |
| 6 |
266032_x_at |
AT2G05890
|
unknown |
0.544 |
-0.336 |
| 7 |
253827_at |
AT4G28085
|
unknown |
0.540 |
-0.066 |
| 8 |
254205_at |
AT4G24170
|
[ATP binding microtubule motor family protein] |
0.521 |
-0.210 |
| 9 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
0.514 |
-0.362 |
| 10 |
252858_at |
AT4G39770
|
TPPH, trehalose-6-phosphate phosphatase H |
0.509 |
-0.325 |
| 11 |
264534_at |
AT1G55700
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.502 |
-0.368 |
| 12 |
247704_at |
AT5G59510
|
DVL18, DEVIL 18, RTFL5, ROTUNDIFOLIA like 5 |
0.479 |
-0.007 |
| 13 |
250327_at |
AT5G12050
|
unknown |
0.479 |
-0.444 |
| 14 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
0.472 |
-0.288 |
| 15 |
255197_x_at |
AT4G07450
|
unknown |
0.450 |
-0.251 |
| 16 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.446 |
-0.281 |
| 17 |
249893_at |
AT5G22555
|
unknown |
0.443 |
-0.365 |
| 18 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
0.442 |
-0.459 |
| 19 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.442 |
-0.364 |
| 20 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.441 |
-0.337 |
| 21 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.429 |
-0.470 |
| 22 |
246075_at |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
0.425 |
-0.838 |
| 23 |
248411_at |
AT5G51580
|
unknown |
0.423 |
-0.239 |
| 24 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.396 |
-0.124 |
| 25 |
251238_at |
AT3G62430
|
[Protein with RNI-like/FBD-like domains] |
0.386 |
-0.101 |
| 26 |
253179_at |
AT4G35200
|
unknown |
0.369 |
-0.438 |
| 27 |
253133_at |
AT4G35800
|
NRPB1, RNA polymerase II large subunit, RNA_POL_II_LS, RNA POLYMERASE II LARGE SUBUNIT, RNA_POL_II_LSRNA_POL_II_LS, RPB1, RNA POLYMERASE II LARGE SUBUNIT |
0.338 |
-0.334 |
| 28 |
258307_x_at |
AT3G30560
|
[similar to AT hook motif-containing protein-related [Arabidopsis thaliana] (TAIR:AT1G35940.1)] |
0.333 |
-0.264 |
| 29 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.331 |
-0.007 |
| 30 |
263837_at |
AT2G04500
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.325 |
-0.182 |
| 31 |
261142_at |
AT1G19780
|
ATCNGC8, cyclic nucleotide gated channel 8, CNGC8, cyclic nucleotide gated channel 8 |
0.313 |
-0.146 |
| 32 |
263070_at |
AT2G17600
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.310 |
-0.197 |
| 33 |
260103_at |
AT1G35430
|
unknown |
0.303 |
-0.091 |
| 34 |
259481_at |
AT1G18970
|
GLP4, germin-like protein 4 |
0.300 |
-0.057 |
| 35 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
0.297 |
-0.645 |
| 36 |
254193_at |
AT4G23870
|
unknown |
0.296 |
-0.697 |
| 37 |
253814_at |
AT4G28290
|
unknown |
0.294 |
0.104 |
| 38 |
253318_at |
AT4G33970
|
[Leucine-rich repeat (LRR) family protein] |
0.293 |
-0.033 |
| 39 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
0.291 |
-0.300 |
| 40 |
250958_at |
AT5G03260
|
LAC11, laccase 11 |
0.288 |
0.049 |
| 41 |
247137_at |
AT5G66210
|
CPK28, calcium-dependent protein kinase 28 |
0.284 |
-0.230 |
| 42 |
248358_at |
AT5G52400
|
CYP715A1, cytochrome P450, family 715, subfamily A, polypeptide 1 |
0.281 |
0.335 |
| 43 |
259738_at |
AT1G64355
|
unknown |
0.273 |
-0.090 |
| 44 |
247751_at |
AT5G59050
|
unknown |
0.273 |
-0.108 |
| 45 |
257511_at |
AT1G43000
|
[PLATZ transcription factor family protein] |
0.268 |
-0.112 |
| 46 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.268 |
-0.221 |
| 47 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
0.266 |
0.086 |
| 48 |
260030_at |
AT1G68880
|
AtbZIP, basic leucine-zipper 8, bZIP, basic leucine-zipper 8 |
-0.988 |
0.853 |
| 49 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.723 |
0.752 |
| 50 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.667 |
0.824 |
| 51 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-0.664 |
0.544 |
| 52 |
246215_at |
AT4G37180
|
[Homeodomain-like superfamily protein] |
-0.664 |
0.709 |
| 53 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.592 |
0.500 |
| 54 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.562 |
0.523 |
| 55 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
-0.558 |
0.510 |
| 56 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
-0.532 |
0.267 |
| 57 |
251956_at |
AT3G53460
|
CP29, chloroplast RNA-binding protein 29 |
-0.523 |
0.535 |
| 58 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
-0.511 |
0.436 |
| 59 |
262876_at |
AT1G64750
|
ATDSS1(I), deletion of SUV3 suppressor 1(I), DSS1(I), deletion of SUV3 suppressor 1(I) |
-0.497 |
0.086 |
| 60 |
255586_at |
AT4G01560
|
MEE49, maternal effect embryo arrest 49 |
-0.490 |
0.404 |
| 61 |
265966_at |
AT2G37220
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.486 |
0.023 |
| 62 |
264188_at |
AT1G54690
|
G-H2AX, GAMMA H2AX, GAMMA-H2AX, gamma histone variant H2AX, H2AXB, HTA3, histone H2A 3 |
-0.463 |
0.355 |
| 63 |
256978_at |
AT3G21110
|
ATPURC, PUR7, purin 7, PURC, PURIN C |
-0.461 |
0.370 |
| 64 |
258155_at |
AT3G18130
|
RACK1C, receptor for activated C kinase 1C, RACK1C_AT, receptor for activated C kinase 1C |
-0.461 |
0.296 |
| 65 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
-0.460 |
0.372 |
| 66 |
247912_at |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.449 |
0.258 |
| 67 |
267102_at |
AT2G41500
|
EMB2776, LIS, LACHESIS |
-0.427 |
0.230 |
| 68 |
258794_at |
AT3G04710
|
TPR10, tetratricopeptide repeat 10 |
-0.420 |
0.319 |
| 69 |
251740_at |
AT3G56070
|
ROC2, rotamase cyclophilin 2 |
-0.417 |
0.421 |
| 70 |
254037_at |
AT4G25760
|
ATGDU2, glutamine dumper 2, GDU2, glutamine dumper 2 |
-0.413 |
0.303 |
| 71 |
261911_at |
AT1G80750
|
[Ribosomal protein L30/L7 family protein] |
-0.397 |
0.388 |
| 72 |
266980_at |
AT2G39390
|
[Ribosomal L29 family protein ] |
-0.394 |
0.311 |
| 73 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.393 |
0.407 |
| 74 |
246278_at |
AT4G37190
|
[LOCATED IN: cytosol, plasma membrane] |
-0.386 |
0.288 |
| 75 |
245122_at |
AT2G47420
|
DIM1A, adenosine dimethyl transferase 1A |
-0.378 |
0.296 |
| 76 |
262415_at |
AT1G49400
|
emb1129, embryo defective 1129 |
-0.376 |
0.347 |
| 77 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.368 |
0.486 |
| 78 |
248254_at |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-0.367 |
-0.087 |
| 79 |
261655_at |
AT1G01940
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.366 |
0.344 |
| 80 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.365 |
0.556 |
| 81 |
250222_at |
AT5G14050
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.362 |
0.255 |
| 82 |
249466_at |
AT5G39740
|
OLI7, OLIGOCELLULA 7, RPL5B, ribosomal protein L5 B |
-0.362 |
0.034 |
| 83 |
253687_at |
AT4G29520
|
[LOCATED IN: endoplasmic reticulum, plasma membrane] |
-0.361 |
0.096 |
| 84 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
-0.360 |
0.284 |
| 85 |
249938_at |
AT5G22330
|
ATTIP49A, RIN1, RESISTANCE TO PSEUDOMONAS SYRINGAE PV MACULICOLA INTERACTOR 1 |
-0.360 |
0.286 |
| 86 |
259908_at |
AT1G60850
|
AAC42, ATRPAC42 |
-0.354 |
0.255 |
| 87 |
249901_at |
AT5G22650
|
ATHD2, ARABIDOPSIS HISTONE DEACETYLASE 2, ATHD2B, HD2, HISTONE DEACETYLASE 2, HD2B, histone deacetylase 2B, HDA4, HDT02, HDT2 |
-0.354 |
0.208 |
| 88 |
257018_at |
AT3G19630
|
[Radical SAM superfamily protein] |
-0.353 |
0.040 |
| 89 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
-0.352 |
0.276 |
| 90 |
254684_at |
AT4G13850
|
ATGRP2, GLYCINE-RICH RNA-BINDING PROTEIN 2, GR-RBP2, glycine-rich RNA-binding protein 2, GRP2, glycine rich protein 2 |
-0.348 |
0.182 |
| 91 |
256173_at |
AT1G51730
|
[Ubiquitin-conjugating enzyme family protein] |
-0.344 |
0.133 |
| 92 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
-0.344 |
0.322 |
| 93 |
251638_at |
AT3G57490
|
[Ribosomal protein S5 family protein] |
-0.337 |
0.255 |