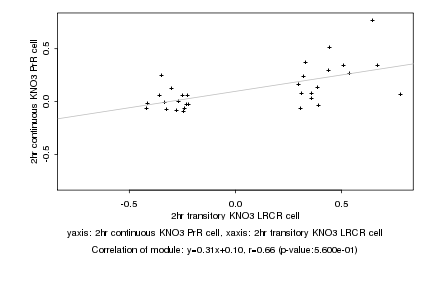
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr transitory KNO3 LRCR cell |
Log2 signal ratio
2hr continuous KNO3 PrR cell |
| 1 |
257810_at |
AT3G27070
|
TOM20-1, translocase outer membrane 20-1 |
0.776 |
0.070 |
| 2 |
261630_at |
AT1G50080
|
unknown |
0.666 |
0.347 |
| 3 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.643 |
0.773 |
| 4 |
252196_at |
AT3G50200
|
unknown |
0.533 |
0.274 |
| 5 |
252191_at |
AT3G50180
|
unknown |
0.506 |
0.344 |
| 6 |
262399_at |
AT1G49500
|
unknown |
0.441 |
0.515 |
| 7 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
0.438 |
0.300 |
| 8 |
267099_at |
AT2G41460
|
ARP, apurinic endonuclease-redox protein |
0.388 |
-0.034 |
| 9 |
253587_at |
AT4G30770
|
[Putative membrane lipoprotein] |
0.383 |
0.136 |
| 10 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.358 |
0.081 |
| 11 |
266889_at |
AT2G44640
|
unknown |
0.355 |
0.035 |
| 12 |
245117_at |
AT2G41560
|
ACA4, autoinhibited Ca(2+)-ATPase, isoform 4 |
0.329 |
0.371 |
| 13 |
247149_at |
AT5G65660
|
[hydroxyproline-rich glycoprotein family protein] |
0.319 |
0.244 |
| 14 |
263775_at |
AT2G46410
|
CPC, CAPRICE |
0.311 |
0.083 |
| 15 |
249961_at |
AT5G18770
|
[F-box/FBD-like domains containing protein] |
0.303 |
-0.064 |
| 16 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.297 |
0.165 |
| 17 |
252206_at |
AT3G50360
|
ATCEN2, centrin2, CEN1, CENTRIN 1, CEN2, centrin2 |
-0.422 |
-0.060 |
| 18 |
246459_at |
AT5G16900
|
[Leucine-rich repeat protein kinase family protein] |
-0.419 |
-0.015 |
| 19 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.360 |
0.060 |
| 20 |
244933_at |
ATCG01070
|
NDHE |
-0.350 |
0.253 |
| 21 |
260764_at |
AT1G48950
|
[C3HC zinc finger-like ] |
-0.338 |
-0.004 |
| 22 |
254209_at |
AT4G23490
|
unknown |
-0.327 |
-0.073 |
| 23 |
258339_at |
AT3G16120
|
[Dynein light chain type 1 family protein] |
-0.304 |
0.125 |
| 24 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
-0.279 |
-0.079 |
| 25 |
260009_at |
AT1G67950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.272 |
0.003 |
| 26 |
249471_at |
AT5G39360
|
EDL2, EID1-like 2 |
-0.250 |
0.061 |
| 27 |
248501_at |
AT5G50440
|
ATMEMB12, MEMB12, membrin 12 |
-0.245 |
-0.089 |
| 28 |
258929_at |
AT3G10060
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.244 |
-0.057 |
| 29 |
262965_at |
AT1G54310
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.233 |
-0.020 |
| 30 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
-0.228 |
0.058 |
| 31 |
259497_at |
AT1G15860
|
unknown |
-0.222 |
-0.027 |