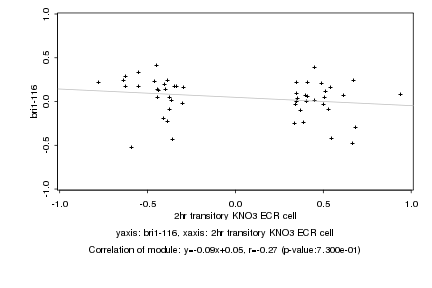
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr transitory KNO3 ECR cell |
Log2 signal ratio
bri1-116 |
| 1 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.937 |
0.091 |
| 2 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.680 |
-0.294 |
| 3 |
262912_at |
AT1G59740
|
AtNPF4.3, NPF4.3, NRT1/ PTR family 4.3 |
0.668 |
0.247 |
| 4 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
0.661 |
-0.469 |
| 5 |
266700_at |
AT2G19740
|
[Ribosomal protein L31e family protein] |
0.610 |
0.078 |
| 6 |
246142_at |
AT5G19970
|
unknown |
0.542 |
-0.416 |
| 7 |
261791_at |
AT1G16170
|
unknown |
0.539 |
0.163 |
| 8 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.525 |
-0.089 |
| 9 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
0.512 |
0.124 |
| 10 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.506 |
0.048 |
| 11 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
0.501 |
-0.031 |
| 12 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.488 |
0.206 |
| 13 |
249266_at |
AT5G41670
|
[6-phosphogluconate dehydrogenase family protein] |
0.446 |
0.021 |
| 14 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.445 |
0.396 |
| 15 |
247218_at |
AT5G65010
|
ASN2, asparagine synthetase 2 |
0.409 |
0.226 |
| 16 |
247443_at |
AT5G62720
|
[Integral membrane HPP family protein] |
0.407 |
0.064 |
| 17 |
262180_at |
AT1G78050
|
PGM, phosphoglycerate/bisphosphoglycerate mutase |
0.402 |
0.010 |
| 18 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.397 |
0.079 |
| 19 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.384 |
-0.237 |
| 20 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.368 |
-0.094 |
| 21 |
264760_at |
AT1G61290
|
ATSYP124, SYP124, syntaxin of plants 124 |
0.352 |
0.042 |
| 22 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.347 |
0.002 |
| 23 |
259015_at |
AT3G07350
|
unknown |
0.342 |
0.223 |
| 24 |
251396_at |
AT3G60750
|
AtTKL1, TKL1, transketolase 1 |
0.342 |
0.092 |
| 25 |
247775_at |
AT5G58690
|
ATPLC5, PLC5, phosphatidylinositol-speciwc phospholipase C5 |
0.339 |
-0.027 |
| 26 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
0.335 |
-0.250 |
| 27 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
-0.782 |
0.220 |
| 28 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.638 |
0.248 |
| 29 |
264636_at |
AT1G65490
|
unknown |
-0.631 |
0.172 |
| 30 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
-0.630 |
0.295 |
| 31 |
249037_at |
AT5G44130
|
FLA13, FASCICLIN-like arabinogalactan protein 13 precursor |
-0.592 |
-0.519 |
| 32 |
264837_at |
AT1G03600
|
PSB27 |
-0.557 |
0.182 |
| 33 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.556 |
0.335 |
| 34 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
-0.461 |
0.230 |
| 35 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.451 |
0.412 |
| 36 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
-0.446 |
0.051 |
| 37 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.445 |
0.139 |
| 38 |
258993_at |
AT3G08940
|
LHCB4.2, light harvesting complex photosystem II |
-0.441 |
0.129 |
| 39 |
253040_at |
AT4G37800
|
XTH7, xyloglucan endotransglucosylase/hydrolase 7 |
-0.414 |
-0.188 |
| 40 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.409 |
0.202 |
| 41 |
257168_at |
AT3G24430
|
HCF101, HIGH-CHLOROPHYLL-FLUORESCENCE 101 |
-0.403 |
0.140 |
| 42 |
247214_at |
AT5G64850
|
unknown |
-0.390 |
-0.224 |
| 43 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
-0.387 |
0.247 |
| 44 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
-0.380 |
-0.087 |
| 45 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.378 |
0.048 |
| 46 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.366 |
0.022 |
| 47 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.363 |
-0.424 |
| 48 |
248961_at |
AT5G45650
|
[subtilase family protein] |
-0.351 |
0.180 |
| 49 |
265182_at |
AT1G23740
|
AOR, alkenal/one oxidoreductase |
-0.336 |
0.183 |
| 50 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.306 |
-0.022 |
| 51 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.296 |
0.163 |