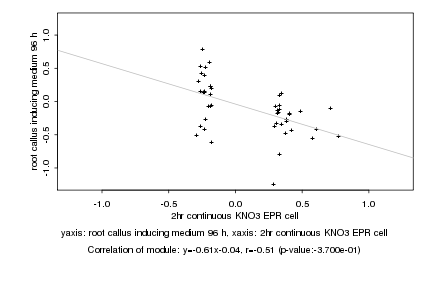
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
2hr continuous KNO3 EPR cell |
Log2 signal ratio
root callus inducing medium 96 h |
| 1 |
257645_at |
AT3G25790
|
[myb-like transcription factor family protein] |
0.769 |
-0.522 |
| 2 |
256045_at |
AT1G07150
|
MAPKKK13, mitogen-activated protein kinase kinase kinase 13 |
0.710 |
-0.104 |
| 3 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.604 |
-0.414 |
| 4 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.572 |
-0.548 |
| 5 |
246142_at |
AT5G19970
|
unknown |
0.484 |
-0.143 |
| 6 |
246854_at |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
0.417 |
-0.435 |
| 7 |
261806_at |
AT1G30510
|
ATRFNR2, root FNR 2, RFNR2, root FNR 2 |
0.403 |
-0.174 |
| 8 |
253179_at |
AT4G35200
|
unknown |
0.400 |
-0.194 |
| 9 |
261791_at |
AT1G16170
|
unknown |
0.381 |
-0.257 |
| 10 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
0.378 |
-0.287 |
| 11 |
262399_at |
AT1G49500
|
unknown |
0.375 |
-0.479 |
| 12 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
0.342 |
0.132 |
| 13 |
263151_at |
AT1G54120
|
unknown |
0.340 |
-0.332 |
| 14 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
0.329 |
-0.054 |
| 15 |
247674_at |
AT5G59930
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.325 |
-0.792 |
| 16 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
0.324 |
-0.106 |
| 17 |
255230_at |
AT4G05390
|
ATRFNR1, root FNR 1, RFNR1, root FNR 1 |
0.324 |
0.103 |
| 18 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.316 |
-0.156 |
| 19 |
266313_at |
AT2G26980
|
CIPK3, CBL-interacting protein kinase 3, SnRK3.17, SNF1-RELATED PROTEIN KINASE 3.17 |
0.312 |
-0.122 |
| 20 |
259727_at |
AT1G60950
|
ATFD2, FERREDOXIN 2, FD2, FED A |
0.309 |
-0.170 |
| 21 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.305 |
-0.325 |
| 22 |
251396_at |
AT3G60750
|
AtTKL1, TKL1, transketolase 1 |
0.294 |
-0.071 |
| 23 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.292 |
-0.364 |
| 24 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.281 |
-1.234 |
| 25 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.298 |
-0.505 |
| 26 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.281 |
0.313 |
| 27 |
256750_at |
AT3G27150
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.267 |
0.533 |
| 28 |
266812_at |
AT2G44830
|
[Protein kinase superfamily protein] |
-0.266 |
0.161 |
| 29 |
260182_at |
AT1G70750
|
MyoB2, myosin binding protein 2 |
-0.264 |
-0.375 |
| 30 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.255 |
0.426 |
| 31 |
262922_at |
AT1G79420
|
unknown |
-0.254 |
0.792 |
| 32 |
245109_at |
AT2G41520
|
DJC62, DNA J protein C62, TPR15, tetratricopeptide repeat 15 |
-0.244 |
0.149 |
| 33 |
251706_at |
AT3G56620
|
UMAMIT10, Usually multiple acids move in and out Transporters 10 |
-0.239 |
-0.418 |
| 34 |
261220_at |
AT1G19970
|
[ER lumen protein retaining receptor family protein] |
-0.238 |
0.403 |
| 35 |
266835_at |
AT2G29990
|
NDA2, alternative NAD(P)H dehydrogenase 2 |
-0.237 |
0.137 |
| 36 |
257715_at |
AT3G12750
|
AtZIP1, ZIP1, zinc transporter 1 precursor |
-0.233 |
0.157 |
| 37 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-0.228 |
0.520 |
| 38 |
266425_at |
AT2G41300
|
SSL1, strictosidine synthase-like 1 |
-0.228 |
-0.270 |
| 39 |
250039_at |
AT5G18370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.207 |
-0.074 |
| 40 |
255727_at |
AT1G25510
|
[Eukaryotic aspartyl protease family protein] |
-0.201 |
0.600 |
| 41 |
248724_at |
AT5G47970
|
[Aldolase-type TIM barrel family protein] |
-0.191 |
0.234 |
| 42 |
266869_at |
AT2G44660
|
[ALG6, ALG8 glycosyltransferase family] |
-0.190 |
0.118 |
| 43 |
261435_at |
AT1G07615
|
[GTP-binding protein Obg/CgtA] |
-0.188 |
0.234 |
| 44 |
256481_at |
AT1G33430
|
[Galactosyltransferase family protein] |
-0.187 |
-0.066 |
| 45 |
264204_at |
AT1G22710
|
ATSUC2, ARABIDOPSIS THALIANA SUCROSE-PROTON SYMPORTER 2, SUC2, sucrose-proton symporter 2, SUT1, SUCROSE TRANSPORTER 1 |
-0.185 |
-0.052 |
| 46 |
249368_at |
AT5G40640
|
unknown |
-0.184 |
0.197 |
| 47 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-0.184 |
-0.614 |