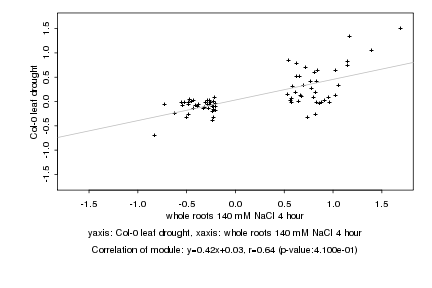
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots 140 mM NaCl 4 hour |
Log2 signal ratio
Col-0 leaf drought |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.687 |
1.517 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.384 |
1.061 |
| 3 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.164 |
1.352 |
| 4 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.147 |
0.751 |
| 5 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
1.144 |
0.842 |
| 6 |
258224_at |
AT3G15670
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.048 |
0.331 |
| 7 |
252073_at |
AT3G51750
|
unknown |
1.022 |
0.128 |
| 8 |
254823_at |
AT4G12580
|
unknown |
1.018 |
0.649 |
| 9 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.957 |
-0.004 |
| 10 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
0.947 |
0.098 |
| 11 |
266274_at |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
0.908 |
0.024 |
| 12 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.875 |
-0.010 |
| 13 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.852 |
-0.024 |
| 14 |
250624_at |
AT5G07330
|
unknown |
0.831 |
0.652 |
| 15 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.823 |
-0.014 |
| 16 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.821 |
0.424 |
| 17 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.819 |
0.205 |
| 18 |
265922_at |
AT2G18480
|
[Major facilitator superfamily protein] |
0.814 |
-0.260 |
| 19 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.805 |
0.617 |
| 20 |
264612_at |
AT1G04560
|
[AWPM-19-like family protein] |
0.795 |
0.097 |
| 21 |
266690_at |
AT2G19900
|
ATNADP-ME1, Arabidopsis thaliana NADP-malic enzyme 1, NADP-ME1, NADP-malic enzyme 1 |
0.770 |
0.278 |
| 22 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.767 |
0.428 |
| 23 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.728 |
-0.311 |
| 24 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.716 |
0.716 |
| 25 |
257672_at |
AT3G20300
|
unknown |
0.697 |
0.347 |
| 26 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.674 |
0.120 |
| 27 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.660 |
0.128 |
| 28 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
0.652 |
0.522 |
| 29 |
246241_at |
AT4G37050
|
AtPLAIVC, phospholipase A IVC, PLA V, PLAIII{beta}, patatin-related phospholipase III beta, PLP4, PATATIN-like protein 4 |
0.636 |
0.016 |
| 30 |
256114_at |
AT1G16850
|
unknown |
0.623 |
0.527 |
| 31 |
260561_at |
AT2G43580
|
[Chitinase family protein] |
0.620 |
0.789 |
| 32 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.606 |
0.199 |
| 33 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.578 |
0.320 |
| 34 |
258695_at |
AT3G09640
|
APX1B, ASCORBATE PEROXIDASE 1B, APX2, ascorbate peroxidase 2 |
0.573 |
-0.001 |
| 35 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
0.568 |
0.073 |
| 36 |
251188_at |
AT3G62730
|
unknown |
0.554 |
0.040 |
| 37 |
263881_at |
AT2G21820
|
unknown |
0.540 |
0.849 |
| 38 |
247120_at |
AT5G65990
|
[Transmembrane amino acid transporter family protein] |
0.532 |
0.145 |
| 39 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.832 |
-0.683 |
| 40 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
-0.735 |
-0.055 |
| 41 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.634 |
-0.240 |
| 42 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.553 |
-0.012 |
| 43 |
257467_at |
AT1G31320
|
LBD4, LOB domain-containing protein 4 |
-0.546 |
-0.068 |
| 44 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-0.524 |
-0.016 |
| 45 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.511 |
-0.322 |
| 46 |
262091_at |
AT1G56160
|
ATMYB72, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 72, MYB72, myb domain protein 72 |
-0.488 |
-0.052 |
| 47 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.485 |
-0.003 |
| 48 |
260753_at |
AT1G49230
|
AtATL78, ATL78, Arabidopsis T?xicos en Levadura 78 |
-0.483 |
-0.266 |
| 49 |
256368_at |
AT1G66800
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.478 |
0.047 |
| 50 |
255608_at |
AT4G01140
|
unknown |
-0.452 |
0.009 |
| 51 |
248723_at |
AT5G47950
|
[HXXXD-type acyl-transferase family protein] |
-0.439 |
0.031 |
| 52 |
249535_at |
AT5G38820
|
[Transmembrane amino acid transporter family protein] |
-0.437 |
-0.137 |
| 53 |
255814_at |
AT1G19900
|
[glyoxal oxidase-related protein] |
-0.416 |
-0.066 |
| 54 |
247116_at |
AT5G65970
|
ATMLO10, MILDEW RESISTANCE LOCUS O 10, MLO10, MILDEW RESISTANCE LOCUS O 10 |
-0.396 |
-0.095 |
| 55 |
262863_at |
AT1G64910
|
[UDP-Glycosyltransferase superfamily protein] |
-0.387 |
-0.047 |
| 56 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.328 |
-0.123 |
| 57 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.317 |
-0.119 |
| 58 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.308 |
-0.002 |
| 59 |
264587_at |
AT1G05200
|
ATGLR3.4, glutamate receptor 3.4, GLR3.4, glutamate receptor 3.4, GLUR3 |
-0.291 |
0.023 |
| 60 |
251154_at |
AT3G63110
|
ATIPT3, isopentenyltransferase 3, IPT3, isopentenyltransferase 3 |
-0.288 |
-0.058 |
| 61 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.286 |
-0.123 |
| 62 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.275 |
-0.051 |
| 63 |
246093_at |
AT5G20550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.267 |
0.027 |
| 64 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.263 |
-0.003 |
| 65 |
246584_at |
AT5G14730
|
unknown |
-0.242 |
-0.205 |
| 66 |
261545_at |
AT1G63530
|
unknown |
-0.241 |
-0.086 |
| 67 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.237 |
-0.383 |
| 68 |
265545_at |
AT2G28250
|
NCRK |
-0.235 |
-0.097 |
| 69 |
251255_at |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.234 |
0.003 |
| 70 |
256025_at |
AT1G58370
|
ATXYN1, ARABIDOPSIS THALIANA XYLANASE 1, RXF12 |
-0.233 |
-0.163 |
| 71 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.232 |
-0.316 |
| 72 |
264191_at |
AT1G54730
|
[Major facilitator superfamily protein] |
-0.219 |
0.088 |
| 73 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.208 |
-0.170 |
| 74 |
261434_at |
AT1G07650
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.207 |
-0.098 |
| 75 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
-0.205 |
-0.034 |