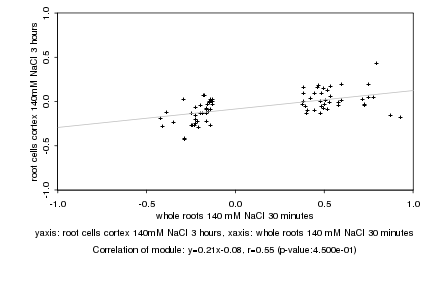
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots 140 mM NaCl 30 minutes |
Log2 signal ratio
root cells cortex 140mM NaCl 3 hours |
| 1 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
0.927 |
-0.170 |
| 2 |
263742_at |
AT2G20625
|
unknown |
0.868 |
-0.157 |
| 3 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.791 |
0.433 |
| 4 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
0.775 |
0.047 |
| 5 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.749 |
0.198 |
| 6 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.745 |
0.051 |
| 7 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.726 |
-0.037 |
| 8 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.724 |
-0.027 |
| 9 |
260744_at |
AT1G15010
|
unknown |
0.715 |
0.024 |
| 10 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.594 |
0.019 |
| 11 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.592 |
0.202 |
| 12 |
245677_at |
AT1G56660
|
unknown |
0.575 |
-0.004 |
| 13 |
245906_at |
AT5G11070
|
unknown |
0.575 |
-0.040 |
| 14 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.535 |
0.179 |
| 15 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.533 |
0.062 |
| 16 |
250098_at |
AT5G17350
|
unknown |
0.526 |
-0.002 |
| 17 |
253643_at |
AT4G29780
|
unknown |
0.518 |
-0.090 |
| 18 |
256442_at |
AT3G10930
|
unknown |
0.517 |
0.127 |
| 19 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.504 |
0.019 |
| 20 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.496 |
-0.026 |
| 21 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.495 |
-0.075 |
| 22 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.491 |
0.156 |
| 23 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.482 |
-0.048 |
| 24 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.479 |
0.092 |
| 25 |
267069_at |
AT2G41010
|
ATCAMBP25, calmodulin (CAM)-binding protein of 25 kDa, CAMBP25, calmodulin (CAM)-binding protein of 25 kDa |
0.478 |
-0.131 |
| 26 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.475 |
0.004 |
| 27 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.466 |
0.183 |
| 28 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.461 |
0.164 |
| 29 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.445 |
0.091 |
| 30 |
258878_at |
AT3G03170
|
unknown |
0.444 |
-0.101 |
| 31 |
247177_at |
AT5G65300
|
unknown |
0.421 |
0.040 |
| 32 |
265732_at |
AT2G01300
|
unknown |
0.403 |
-0.096 |
| 33 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.399 |
-0.132 |
| 34 |
253830_at |
AT4G27652
|
unknown |
0.392 |
-0.046 |
| 35 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.382 |
0.095 |
| 36 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.381 |
0.008 |
| 37 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.379 |
0.163 |
| 38 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.376 |
-0.033 |
| 39 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.425 |
-0.186 |
| 40 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.413 |
-0.277 |
| 41 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.393 |
-0.120 |
| 42 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.351 |
-0.232 |
| 43 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.294 |
0.033 |
| 44 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.290 |
-0.427 |
| 45 |
258540_at |
AT3G06990
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.289 |
-0.413 |
| 46 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.253 |
-0.262 |
| 47 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.249 |
-0.126 |
| 48 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.244 |
-0.270 |
| 49 |
247832_at |
AT5G58550
|
EOL2, ETO1-like 2 |
-0.236 |
-0.264 |
| 50 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.230 |
-0.148 |
| 51 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.227 |
-0.066 |
| 52 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.225 |
-0.245 |
| 53 |
260161_at |
AT1G79860
|
ATROPGEF12, MEE64, MATERNAL EFFECT EMBRYO ARREST 64, ROPGEF12, ROP (rho of plants) guanine nucleotide exchange factor 12 |
-0.225 |
-0.194 |
| 54 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.215 |
-0.225 |
| 55 |
245037_at |
AT2G26420
|
PIP5K3, 1-phosphatidylinositol-4-phosphate 5-kinase 3 |
-0.213 |
-0.291 |
| 56 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.201 |
-0.130 |
| 57 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.199 |
-0.034 |
| 58 |
263226_at |
AT1G30690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.187 |
-0.126 |
| 59 |
255181_at |
AT4G08110
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.2e-66 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.182 |
0.069 |
| 60 |
259134_at |
AT3G05390
|
unknown |
-0.179 |
0.075 |
| 61 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.165 |
-0.215 |
| 62 |
267434_at |
AT2G26260
|
3BETAHSD/D2, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2, AT3BETAHSD/D2, 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 |
-0.165 |
-0.125 |
| 63 |
261505_at |
AT1G71696
|
SOL1, SUPPRESSOR OF LLP1 1 |
-0.165 |
-0.084 |
| 64 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.163 |
-0.071 |
| 65 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.161 |
-0.030 |
| 66 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.157 |
-0.108 |
| 67 |
263085_at |
AT2G16090
|
ARI2, ARIADNE 2, ATARI2, ARABIDOPSIS ARIADNE 2 |
-0.150 |
0.001 |
| 68 |
265056_at |
AT1G51980
|
[Insulinase (Peptidase family M16) protein] |
-0.147 |
-0.011 |
| 69 |
255571_at |
AT4G01220
|
MGP4, male gametophyte defective 4 |
-0.145 |
-0.089 |
| 70 |
263618_at |
AT2G04660
|
APC2, anaphase-promoting complex/cyclosome 2 |
-0.145 |
0.030 |
| 71 |
248916_at |
AT5G45840
|
[Leucine-rich repeat protein kinase family protein] |
-0.141 |
-0.270 |
| 72 |
262868_at |
AT1G64980
|
CDI, cadmium 2+ induced |
-0.135 |
0.009 |
| 73 |
259527_at |
AT1G12600
|
[UDP-N-acetylglucosamine (UAA) transporter family] |
-0.134 |
0.030 |
| 74 |
266322_at |
AT2G46690
|
SAUR32, SMALL AUXIN UPREGULATED RNA 32 |
-0.133 |
-0.025 |
| 75 |
263412_at |
AT2G28720
|
[Histone superfamily protein] |
-0.133 |
0.009 |