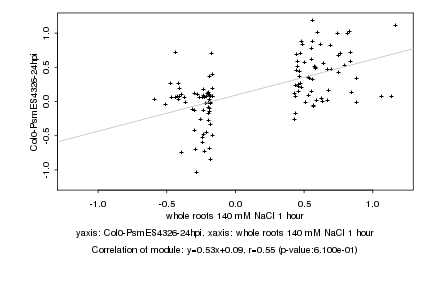
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
whole roots 140 mM NaCl 1 hour |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.163 |
1.114 |
| 2 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.132 |
0.079 |
| 3 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
1.061 |
0.077 |
| 4 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.881 |
0.351 |
| 5 |
263742_at |
AT2G20625
|
unknown |
0.875 |
-0.001 |
| 6 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.842 |
0.143 |
| 7 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.833 |
0.596 |
| 8 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
0.831 |
0.726 |
| 9 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.824 |
1.024 |
| 10 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
0.809 |
1.001 |
| 11 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.794 |
0.539 |
| 12 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.758 |
0.710 |
| 13 |
266841_at |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
0.750 |
0.686 |
| 14 |
260744_at |
AT1G15010
|
unknown |
0.746 |
0.435 |
| 15 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.738 |
1.009 |
| 16 |
251156_at |
AT3G63060
|
EDL3, EID1-like 3 |
0.693 |
0.473 |
| 17 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
0.689 |
0.826 |
| 18 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.672 |
0.163 |
| 19 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.668 |
0.027 |
| 20 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.665 |
0.480 |
| 21 |
251971_at |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
0.636 |
0.556 |
| 22 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.628 |
0.007 |
| 23 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.626 |
0.056 |
| 24 |
265024_at |
AT1G24600
|
unknown |
0.615 |
0.847 |
| 25 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.593 |
1.015 |
| 26 |
264202_at |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
0.583 |
0.020 |
| 27 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.577 |
0.503 |
| 28 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
0.576 |
0.491 |
| 29 |
245677_at |
AT1G56660
|
unknown |
0.572 |
0.519 |
| 30 |
252073_at |
AT3G51750
|
unknown |
0.566 |
-0.063 |
| 31 |
245906_at |
AT5G11070
|
unknown |
0.564 |
-0.046 |
| 32 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.560 |
0.324 |
| 33 |
249197_at |
AT5G42380
|
CML37, calmodulin like 37 |
0.558 |
1.199 |
| 34 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.555 |
0.888 |
| 35 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.550 |
0.779 |
| 36 |
246018_at |
AT5G10695
|
unknown |
0.549 |
0.624 |
| 37 |
251751_at |
AT3G55720
|
unknown |
0.547 |
0.151 |
| 38 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.535 |
0.346 |
| 39 |
247061_at |
AT5G66780
|
unknown |
0.531 |
0.356 |
| 40 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.527 |
0.092 |
| 41 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.504 |
-0.001 |
| 42 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.502 |
0.577 |
| 43 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.487 |
0.841 |
| 44 |
256442_at |
AT3G10930
|
unknown |
0.478 |
0.879 |
| 45 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.474 |
0.218 |
| 46 |
267058_at |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
0.473 |
0.278 |
| 47 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.469 |
0.710 |
| 48 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.464 |
0.370 |
| 49 |
253643_at |
AT4G29780
|
unknown |
0.462 |
0.444 |
| 50 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.459 |
0.158 |
| 51 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.457 |
0.254 |
| 52 |
250098_at |
AT5G17350
|
unknown |
0.453 |
0.224 |
| 53 |
247177_at |
AT5G65300
|
unknown |
0.446 |
0.596 |
| 54 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.445 |
0.524 |
| 55 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.444 |
0.694 |
| 56 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.443 |
0.456 |
| 57 |
258878_at |
AT3G03170
|
unknown |
0.435 |
0.084 |
| 58 |
252321_at |
AT3G48510
|
unknown |
0.434 |
0.248 |
| 59 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.431 |
-0.163 |
| 60 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
0.428 |
0.128 |
| 61 |
248764_at |
AT5G47640
|
NF-YB2, nuclear factor Y, subunit B2 |
0.427 |
-0.253 |
| 62 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.594 |
0.043 |
| 63 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.510 |
-0.029 |
| 64 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.474 |
0.267 |
| 65 |
255608_at |
AT4G01140
|
unknown |
-0.469 |
0.066 |
| 66 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-0.441 |
0.067 |
| 67 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
-0.439 |
0.729 |
| 68 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.426 |
0.080 |
| 69 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.420 |
0.268 |
| 70 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
-0.418 |
0.039 |
| 71 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.414 |
0.199 |
| 72 |
261772_at |
AT1G76240
|
unknown |
-0.411 |
0.075 |
| 73 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.400 |
-0.735 |
| 74 |
259195_at |
AT3G01730
|
unknown |
-0.396 |
0.110 |
| 75 |
252172_at |
AT3G50640
|
unknown |
-0.371 |
0.065 |
| 76 |
249186_at |
AT5G43040
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.364 |
-0.014 |
| 77 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.314 |
-0.110 |
| 78 |
266715_at |
AT2G46780
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.304 |
-0.131 |
| 79 |
245037_at |
AT2G26420
|
PIP5K3, 1-phosphatidylinositol-4-phosphate 5-kinase 3 |
-0.302 |
0.119 |
| 80 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.299 |
-0.417 |
| 81 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.298 |
-0.701 |
| 82 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.284 |
-1.036 |
| 83 |
256157_at |
AT1G13580
|
LAG13, LAG1 longevity assurance homolog 3, LOH3, LAG One Homologue 3 |
-0.282 |
0.109 |
| 84 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.265 |
0.069 |
| 85 |
253227_at |
AT4G35030
|
[Protein kinase superfamily protein] |
-0.261 |
-0.255 |
| 86 |
257172_at |
AT3G23700
|
[Nucleic acid-binding proteins superfamily] |
-0.246 |
-0.598 |
| 87 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.246 |
-0.512 |
| 88 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.240 |
0.065 |
| 89 |
263226_at |
AT1G30690
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.235 |
-0.478 |
| 90 |
250632_at |
AT5G07450
|
CYCP4;3, cyclin p4;3 |
-0.235 |
0.089 |
| 91 |
259888_at |
AT1G76350
|
NLP5, NIN-like protein 5 |
-0.234 |
0.190 |
| 92 |
264114_at |
AT2G31270
|
ATCDT1A, ARABIDOPSIS HOMOLOG OF YEAST CDT1 A, CDT1, ARABIDOPSIS HOMOLOG OF YEAST CDT1, CDT1A, homolog of yeast CDT1 A |
-0.234 |
-0.125 |
| 93 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.231 |
-0.725 |
| 94 |
247024_at |
AT5G66985
|
unknown |
-0.230 |
0.060 |
| 95 |
258832_at |
AT3G07070
|
[Protein kinase superfamily protein] |
-0.225 |
-0.015 |
| 96 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.217 |
0.081 |
| 97 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.213 |
-0.447 |
| 98 |
266109_at |
AT2G37890
|
[Mitochondrial substrate carrier family protein] |
-0.209 |
0.120 |
| 99 |
249031_at |
AT5G44900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.202 |
-0.009 |
| 100 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.202 |
-0.162 |
| 101 |
255576_at |
AT4G01440
|
UMAMIT31, Usually multiple acids move in and out Transporters 31 |
-0.202 |
0.137 |
| 102 |
262868_at |
AT1G64980
|
CDI, cadmium 2+ induced |
-0.198 |
-0.082 |
| 103 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.198 |
-0.264 |
| 104 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
-0.196 |
0.378 |
| 105 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.196 |
0.032 |
| 106 |
260059_at |
AT1G78090
|
ATTPPB, Arabidopsis thaliana trehalose-6-phosphate phosphatase B, TPPB, trehalose-6-phosphate phosphatase B |
-0.195 |
-0.098 |
| 107 |
256275_at |
AT3G12110
|
ACT11, actin-11 |
-0.194 |
-0.675 |
| 108 |
252105_at |
AT3G51470
|
[Protein phosphatase 2C family protein] |
-0.193 |
-0.144 |
| 109 |
265029_at |
AT1G24625
|
ZFP7, zinc finger protein 7 |
-0.189 |
0.104 |
| 110 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.189 |
0.091 |
| 111 |
256360_at |
AT1G66440
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.188 |
0.077 |
| 112 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.185 |
-0.335 |
| 113 |
265205_at |
AT2G36660
|
PAB7, poly(A) binding protein 7 |
-0.182 |
-0.014 |
| 114 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
-0.182 |
-0.834 |
| 115 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.182 |
-0.015 |
| 116 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.180 |
0.708 |
| 117 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
-0.174 |
0.200 |
| 118 |
261505_at |
AT1G71696
|
SOL1, SUPPRESSOR OF LLP1 1 |
-0.173 |
0.081 |
| 119 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.170 |
-0.491 |
| 120 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.168 |
0.075 |
| 121 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
-0.168 |
0.403 |