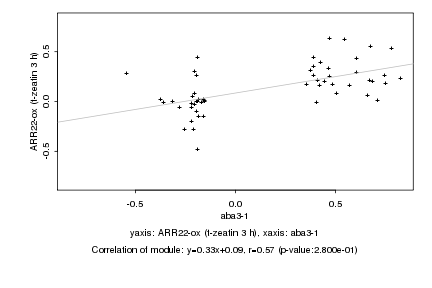
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
aba3-1 |
Log2 signal ratio
ARR22-ox (t-zeatin 3 h) |
| 1 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.823 |
0.232 |
| 2 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.778 |
0.536 |
| 3 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.745 |
0.188 |
| 4 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.743 |
0.269 |
| 5 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.710 |
0.014 |
| 6 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.681 |
0.202 |
| 7 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.672 |
0.553 |
| 8 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.670 |
0.213 |
| 9 |
250476_at |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
0.660 |
0.062 |
| 10 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.603 |
0.439 |
| 11 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.602 |
0.293 |
| 12 |
252977_at |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.569 |
0.164 |
| 13 |
261021_at |
AT1G26380
|
[FAD-binding Berberine family protein] |
0.542 |
0.632 |
| 14 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.501 |
0.083 |
| 15 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.483 |
0.171 |
| 16 |
256989_at |
AT3G28580
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.467 |
0.634 |
| 17 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.467 |
0.253 |
| 18 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.465 |
0.334 |
| 19 |
256874_at |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
0.444 |
0.204 |
| 20 |
256526_at |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
0.424 |
0.401 |
| 21 |
266017_at |
AT2G18690
|
unknown |
0.417 |
0.168 |
| 22 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.408 |
0.218 |
| 23 |
245401_at |
AT4G17670
|
unknown |
0.402 |
-0.004 |
| 24 |
257540_at |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
0.390 |
0.265 |
| 25 |
260015_at |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
0.388 |
0.450 |
| 26 |
262085_at |
AT1G56060
|
unknown |
0.386 |
0.352 |
| 27 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.374 |
0.314 |
| 28 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.355 |
0.178 |
| 29 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.545 |
0.284 |
| 30 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.379 |
0.027 |
| 31 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
-0.360 |
-0.001 |
| 32 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.315 |
0.010 |
| 33 |
249105_at |
AT5G43640
|
[Ribosomal protein S19 family protein] |
-0.281 |
-0.058 |
| 34 |
257672_at |
AT3G20300
|
unknown |
-0.257 |
-0.272 |
| 35 |
251297_at |
AT3G62020
|
GLP10, germin-like protein 10 |
-0.224 |
-0.200 |
| 36 |
265949_at |
AT2G18540
|
[RmlC-like cupins superfamily protein] |
-0.222 |
-0.053 |
| 37 |
250281_at |
AT5G13240
|
[transcription regulators] |
-0.220 |
-0.013 |
| 38 |
254296_at |
AT4G23090
|
unknown |
-0.218 |
0.058 |
| 39 |
249814_at |
AT5G23840
|
[MD-2-related lipid recognition domain-containing protein] |
-0.210 |
-0.274 |
| 40 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.208 |
0.090 |
| 41 |
266611_at |
AT2G14960
|
GH3.1 |
-0.207 |
-0.026 |
| 42 |
255521_at |
AT4G02280
|
ATSUS3, SUS3, sucrose synthase 3 |
-0.207 |
0.311 |
| 43 |
254258_at |
AT4G23410
|
TET5, tetraspanin5 |
-0.197 |
0.264 |
| 44 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
-0.196 |
-0.095 |
| 45 |
266153_at |
AT2G12320
|
unknown |
-0.195 |
0.008 |
| 46 |
255450_at |
AT4G02850
|
[phenazine biosynthesis PhzC/PhzF family protein] |
-0.192 |
-0.476 |
| 47 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.192 |
0.449 |
| 48 |
263617_at |
AT2G04670
|
[gypsy-like retrotransposon family, has a 1.2e-313 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-0.190 |
0.003 |
| 49 |
256444_at |
AT3G11060
|
unknown |
-0.189 |
0.030 |
| 50 |
255005_at |
AT4G09990
|
GXM2, glucuronoxylan methyltransferase 2 |
-0.186 |
-0.142 |
| 51 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
-0.173 |
-0.001 |
| 52 |
247633_at |
AT5G60470
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.164 |
0.030 |
| 53 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.160 |
-0.147 |
| 54 |
252243_at |
AT3G50120
|
unknown |
-0.158 |
0.012 |
| 55 |
252460_at |
AT3G47230
|
unknown |
-0.157 |
0.006 |